Monitoring protein folding through high pressure NMR spectroscopy
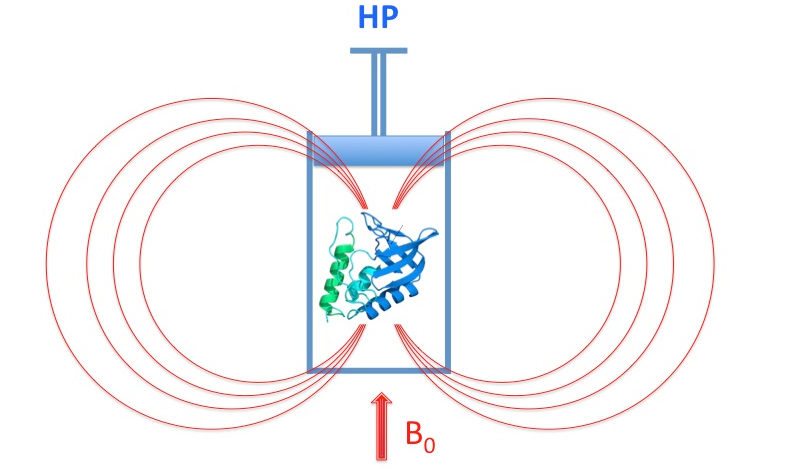
Everything you always wanted to know on the use of High-Pressure NMR to study Protein Folding (but were afraid to ask).

Everything you always wanted to know on the use of High-Pressure NMR to study Protein Folding (but were afraid to ask).

HOW TO GET 3D RECONSTRUCTION OF SUPRAMOLECULAR ASSEMBLIES FROM 2D SUPER-RESOLUTION LIGHT MICROSCOPY IMAGES?
Super-resolution microscopies enable imaging of supramolecular assemblies in complex environments or in live cells with chemical specificity at spatial resolutions surpassing the diffraction limit. A main limitation remains the statistical analysis of individual images to dissect heterogeneity, and to increase image fidelity and resolution.
Today, the team of Marcelo Nollmann (CBS, Montpellier) and Gaëtan Bellot (IGF, Team. Lebon) describe a single-particle reconstruction method that enables template-free reconstruction of nanomolecular structures from two-dimensional super-resolution projection images to achieve isotropic three-dimensional resolutions.
This research has been recently highlighted in Nature Methods (Nature Methods 14, 942 (2017) doi:10.1038/nmeth.4453).

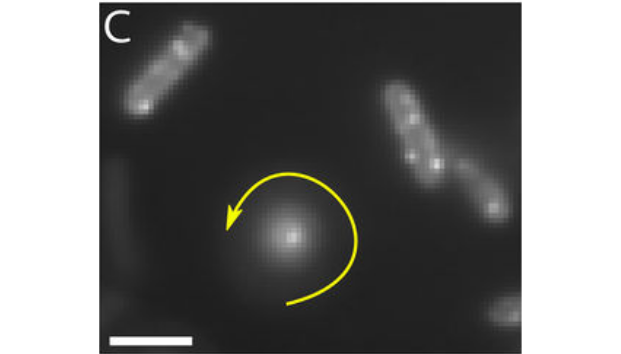
The bacterial flagellar motor (BFM) is the rotary motor that rotates each bacterial flagellum, powering the swimming and swarming of many motile bacteria. The torque is provided by stator units, ion motive force-powered ion channels known to assemble and disassemble dynamically in the BFM. This turnover is mechanosensitive, with the number of engaged units dependent on the viscous load experienced by the motor through the flagellum.
In this work, we directly measure the kinetics of arrival and departure of the stator units in individual motors via analysis of high-resolution recordings of motor speed, while dynamically varying the load on the motor via external magnetic torque.
The kinetic rates obtained, robust with respect to the details of the applied adsorption model, indicate that the lifetime of an assembled stator unit increases when a higher force is applied to its anchoring point in the cell wall. This provides strong evidence that a catch bond (a bond strengthened instead of weakened by force) drives mechanosensitivity of the flagellar motor complex.
These results add the BFM to a short, but growing, list of systems demonstrating catch bonds, suggesting that this “molecular strategy” is a widespread mechanism to sense and respond to mechanical stress. We propose that force-enhanced stator adhesion allows the cell to adapt to a heterogeneous environmental viscosity and may ultimately play a role in surface-sensing during swarming and biofilm formation.
Fluorescent fusion proteins open a direct and unique window onto protein function. However, they also introduce the risk of perturbation of the function of the native protein. Successful applications of fluorescent fusions therefore rely on a careful assessment and minimization of the side effects, but such insight is still lacking for many applications. This is particularly relevant in the study of the internal dynamics of motor proteins, where both the chemical and mechanical reaction coordinates can be affected. Fluorescent proteins fused to the stator of the Bacterial Flagellar Motor (BFM) have previously been used to unveil the motor subunit dynamics. Here we report the effects on single motors of three fluorescent proteins fused to the stators, all of which altered BFM behavior.
Scientific Reports 7, Article number: 12583(2017)
doi:10.1038/s41598-017-11241-w
https://www.nature.com/articles/s41598-017-11241-w
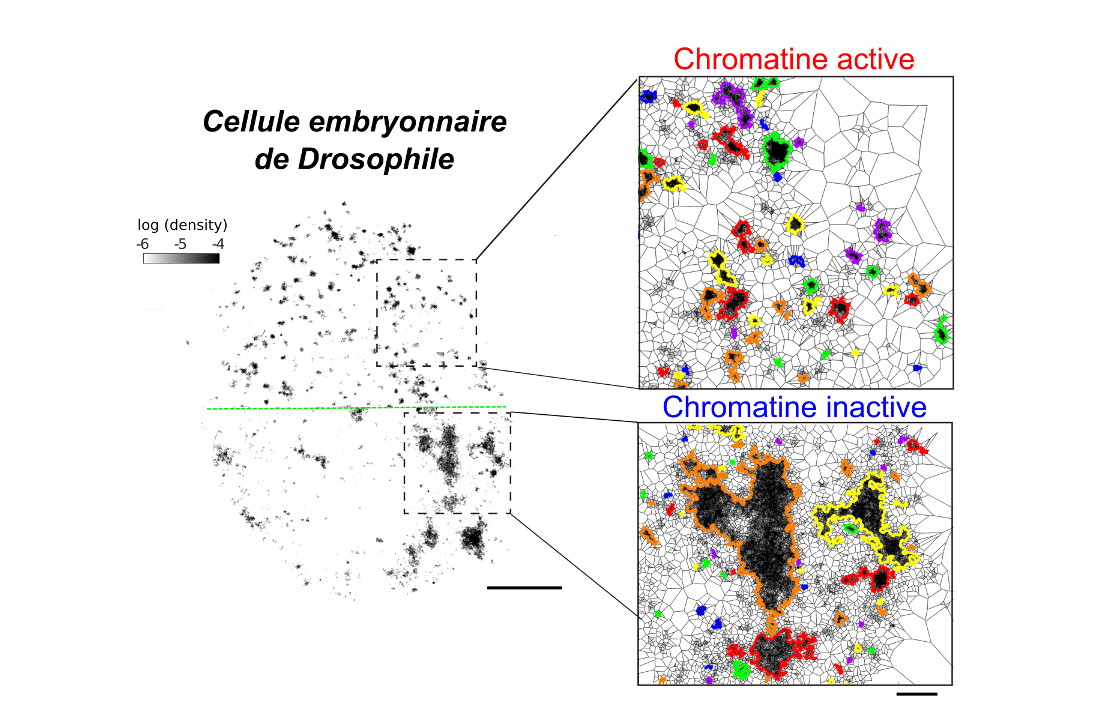
The organization of DNA in the three-dimensional space of the cell nucleus determines the regulation of most cellular activities. In recent years, a new level of organization has been discovered and its role in different pathologies has confirmed its critical importance. This level of organization rises from the preferential interaction between certain parts of the DNA and the exclusion of others. The mechanisms governing the assembly of these structures were still poorly understood.
In this study we labeled dozens of specific regions of DNA and quantified their interaction frequency using three-dimensional super-resolution microscopy. We have studied hundreds of Drosophila embryonic cells in different states of development and found that the frequency of each of these contacts changes according to cell type and its metabolic state. Next, we assessed whether these changes in interaction frequency were reflected at a larger scale of DNA organization by fluorescently labeling epigenetic markers of active and inactive chromatin.
From single molecule clusterization analysis, we have shown that DNA forms nano-compartments with distinct characteristics depending on whether chromatin is active or inactive and that the number and size of these compartments changes between different cell types. These results show that the regulation of the interaction frequency between specific regions plays a key role in the organization of chromatin at several scales. Other studies could reveal how alterations of these interactions could create pathological manifestations in the cell and also affect the development of a complete organism during embryogenesis.
This research has been recently highlighted in Nature Communications (Nature Communications, doi:10.1038/s41467-017-01962-x).