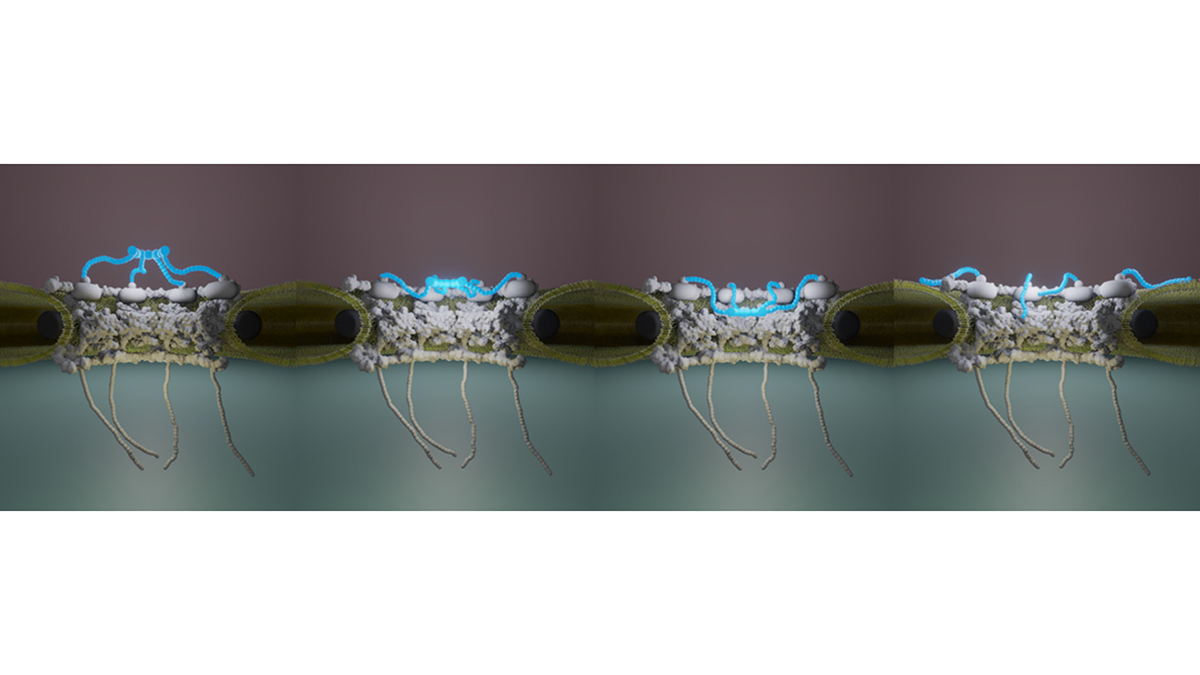
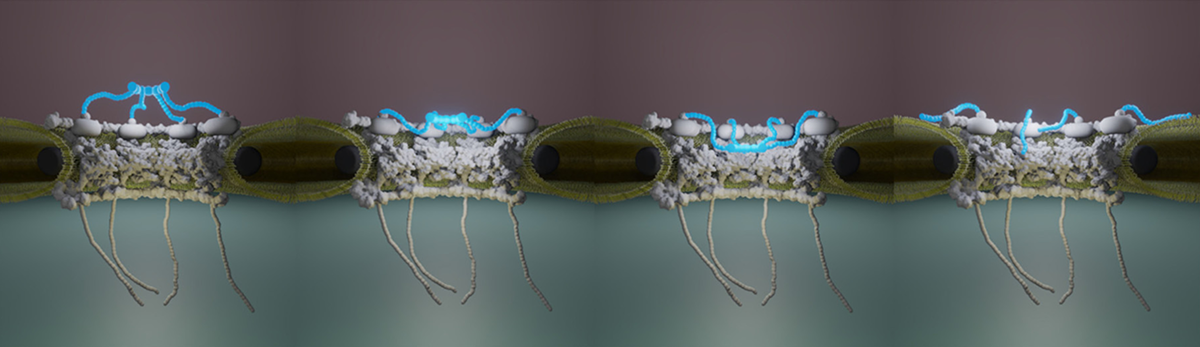
Multi-scale dynamic imaging reveals that cooperative motility behaviors promote efficient predation in bacteria
Overlay of Myxococcus xanthus bacteria trajectories
during the invasion of an Escherichia coli colony.
Publication link:
Rombouts, S., Mas, A., Le Gall, A. Fiche, J.B., Mignot, T., Nollmann, M.. Multi-scale dynamic imaging reveals that cooperative motility behaviors promote efficient predation in bacteria. Nat Commun 14, 5588 (2023). https://doi.org/10.1038/s41467-023-41193-x
Video: “Synchronized synergy: collective movement in Myxococcus xanthus”
https://www.wibbitz.com/watch/b603898e8c7d54592955b58828cb8d349/?cl=#5c6168&cl4=%2315324e&lg=8132e369779d4f878702ab25f531c8cf&type=produced
Abstract
Many species, such as fish schools or bird flocks, rely on collective motion to forage, prey, or escape predators. Likewise, Myxococcus xanthus forages and moves collectively to prey and feed on other bacterial species. These activities require two distinct motility machines enabling adventurous (A) and social (S) gliding, however when and how these mechanisms are used has remained elusive. Here, we address this long-standing question by applying multiscale semantic cell tracking during predation. We show that: (1) foragers and swarms can comprise A- and S-motile cells, with single cells exchanging frequently between these groups; (2) A-motility is critical to ensure the directional movement of both foragers and swarms; (3) the combined action of A- and S-motile cells within swarms leads to increased predation efficiencies. These results challenge the notion that A- and S-motilities are exclusive to foragers and swarms, and show that these machines act synergistically to enhance predation efficiency.
2D DNA-origami assembly for Cryo-EM Applications
We propose a self-assembled DNA origami honeycomb 2D-lattice as a molecular imaging scaffold for Cryo-Electron Microscopy (Cryo-EM). The thin, micrometer-scale supporting scaffold is able to cover extended areas of the holey carbon film and sufficiently resilient to withstand blotting and plunge-freezing forces. Furthermore, the DNA binding sites can be chemically engineered for selective surface-affinity trapping. We demonstrate the advantages of the method to facilitate membrane vesicles sample preparation. This includes increasing the local density of the vesicles, enabling the study of low-abundance membrane complexes, and purifying heterogenous samples by isolating vesicles from cell conditioned medium.
Free-Standing DNA Origami Superlattice to Facilitate Cryo-EM Visualization of Membrane Vesicles
Nesrine Aissaoui*, Allan Mills*, Josephine Lai-Kee-Him*, Nicolas Triomphe, Quentin Cece, Christine Doucet, Anne Bonhoure, Michel Vidal, Yonggang Ke, and Gaetan Bellot.