Transcription and chromosome observation in single cells with Hi-M
Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M
Andrés M. Cardozo Gizzi, Sergio M. Espinola, Julian Gurgo, Christophe Houbron, Jean-Bernard Fiche, Diego I. Cattoni, Marcelo Nollmann
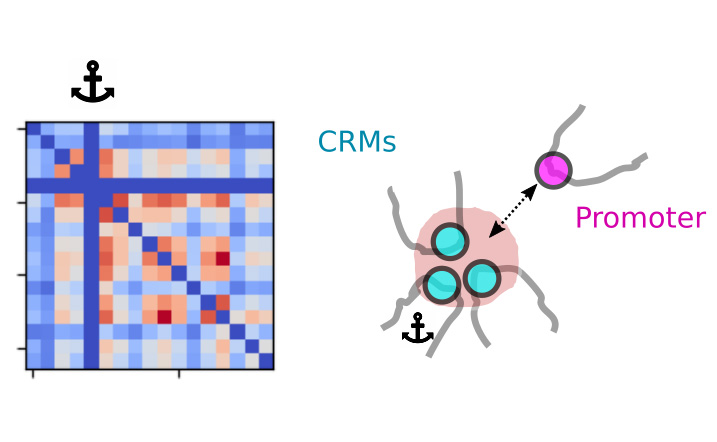
Simultaneous observation of 3D chromatin organization and transcription at the single cell level and with high spatial resolution may hold the key to unveil the mechanisms regulating embryonic development, cell differentiation and even disease. We have recently developed Hi-M, a technology that allows for the sequential labelling, 3D imaging and localization of multiple genomic DNA loci together with RNA expression in single cells within whole, intact Drosophila embryos. Importantly, Hi-M enables simultaneous detection of RNA expression and chromosome organization without requiring sample unmounting and primary probe re-hybridization. Here, we provide a step-by-step protocol describing the design of probes, the preparation of samples, the stable immobilization of embryos into microfluidics chambers, and the complete procedure for image acquisition. The combined RNA/DNA fluorescence in situ hybridization procedure takes 4-5 days including embryo collection. In addition, we describe image analysis software to segment nuclei, detect genomic spots, correct for drift and produce Hi-M matrices. A typical Hi-M experiment takes 1-2 days to complete all rounds of labelling and imaging and 4 additional days for image analysis. This technology can be easily expanded to investigate cell differentiation in cultured cells, or organization of chromatin within complex tissues.
Nat Protoc 2020 Mar;15(3):840-876. doi: 10.1038/s41596-019-0269-9
ATP-driven separation of liquid phase condensates in bacteria
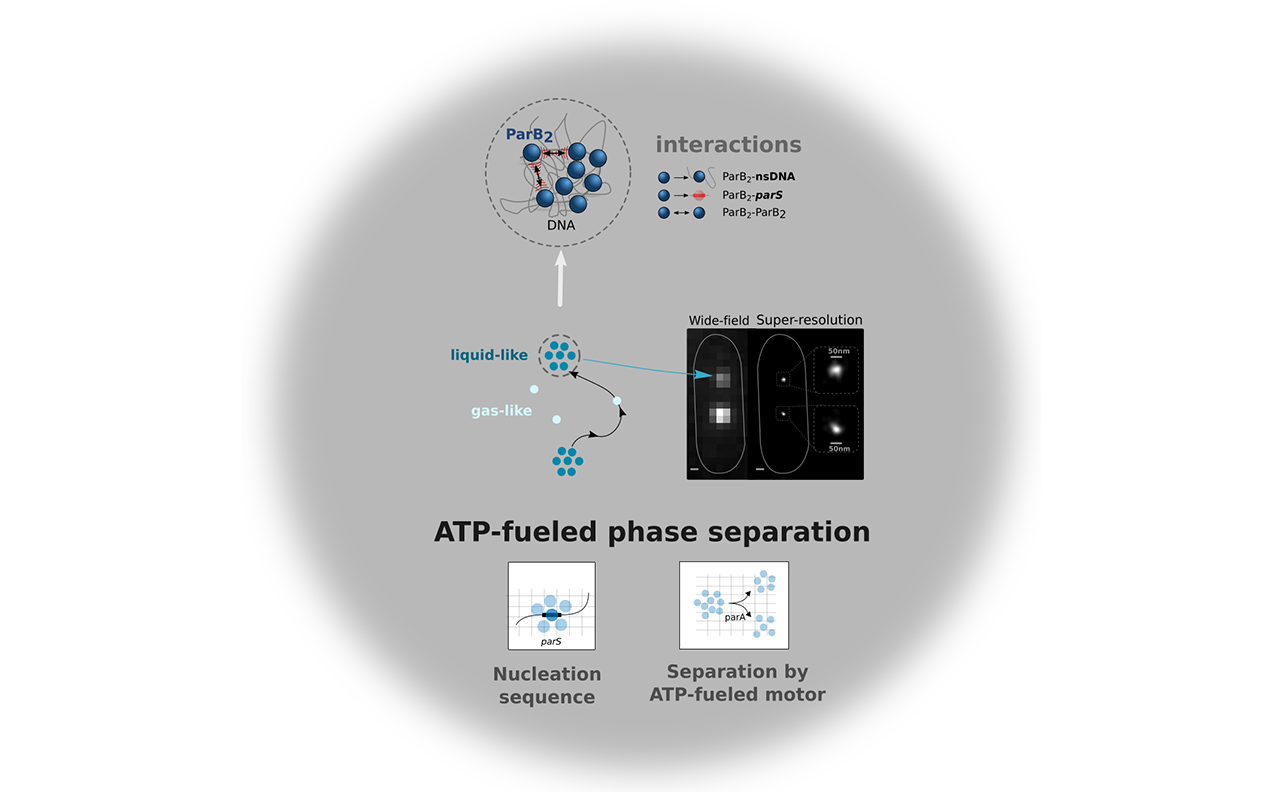
Liquid-liquid phase separated (LLPS) states are key to compartmentalise components in the absence of membranes, however it is unclear whether LLPS condensates are actively and specifically organized in the sub-cellular space and by which mechanisms. Here, we address this question by focusing on the ParABS DNA segregation system, composed of a centromeric-like sequence (parS), a DNA-binding protein (ParB) and a motor (ParA). We show that parS-ParB associate to form nanometer-sized, round condensates. ParB molecules diffuse rapidly within the nucleoid volume, but display confined motions when trapped inside ParB condensates. Single ParB molecules are able to rapidly diffuse between different condensates, and nucleation is strongly favoured by parS. Notably, the ParA motor is required to prevent the fusion of ParB condensates. These results describe a novel active mechanism that splits, segregates and localises non-canonical LLPS condensates in the sub-cellular space.
B. Guilhas, J.C. Walter, J. Rech, G. David, N.-O. Walliser, J. Palmeri, C., Mathieu-Demaziere, A. Parmeggiani, J.Y. Bouet, A. Le Gall1, M. Nollmann
Full text: https://doi.org/10.1101/791368
CNRS news: https://insb.cnrs.fr/fr/cnrsinfo/comment-la-separation-de-phases-aide-la-segregation-du-materiel-genetique