A. Padilla, K. de Guillen, N. Declerck, L. Mammri
An alternative strategy to pesticides for crop protection is the breeding of resistant plant varieties. Key elements of plant resistance to infections are the immune receptors (IRs) of the NLR type that recognize pathogen-secreted virulence factors, also termed avirulence (AVR) effectors. Our project aims at understanding the role and evolution of host/pathogen resistance systems by analyzing protein structures, mode of interaction and allelic diversity within families of plant NLR receptors and AVR effectors secreted by fungi responsible of severe cereal diseases.
 |
 |
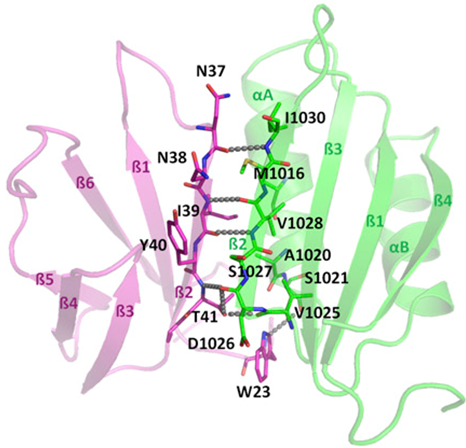 |
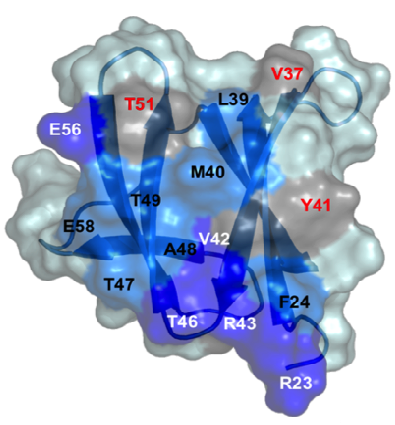
By combining structural and bio-informatic approaches we discovered a large and diverse family of structurally conserved virulence factors, the MAX effectors originally identified in the rice blast fungus Magnaporthe oryzae. Some MAX effectors are specifically recognized by NLR-type immuno receptors but the cellular mechanisms they target in cereals are largely unknown. In a collaborative work with INRA we focused on the RGA4-RGA5 pair of IRs from rice and we elucidated the 3D structure of the RGA5HMA (sensor domain) in complex with the AVR1-CO39 or AVR-Pia effectors from M. oryzae. Structure-based mutational analyses have been performed to validate the RGA5HMA/AVR interaction surface and design new recognition specificities. We now seek to determine the structure of other predicted MAX effectors by NMR and of the full-length RGA4 or RGA5 receptors by crystallography and/or cryo-electron microscopy. In vitro and in vivo characterization of the proteins and their interactions are also in progress using state-of-the-art fluorescence microscopy and spectroscopy approaches.
Collaboration: T. Kroj and S. Cesari (BGPI, INRA Montpellier)
References : deGuillen et al., PLoS Pathog, 2015; Ortiz, deGuillen et al., Plant Cell, 2017; Guo et al., Proc Natl Acad Sci USA, 2018

