VITO2

An interactive online or local Java tool for homology modelling and protein family study coupling a full featured alignment editor with a complete 3D viewer.
Grand F, Pons JL, Labesse, G (2016)
Lahfa M, Barthe P, deGuillen K, Cesari S, Raji M, Kroj T, Le Naour-Vernet M, Hoh F, Gladieux P, Roumestand C, Gracy J, Declerck N, Padilla A, PLoS Pathog, 2024
Aissaoui N, Mills A, Lai-Kee-Him J, Triomphe N, Cece Q, Doucet C, Bonhoure A, Vidal M, Ke Y, Bellot G, J Am Chem Soc, 2024
Gurgo J, Walter JC, Fiche JB, Houbron C, Schaeffer M, Cavalli G, Bantignies F, Nollmann M, Cell Rep, 2024
Varela Salgado M, Adriaans IE, Touati SA, Ibanes S, Lai-Kee-Him J, Ancelin A, Cipelletti L, Picas L, Piatti S, Nat Commun, 2024
Mouhand A, Nakatani K, Kono F, Hippo Y, Matsuo T, Barthe P, Peters J, Suenaga Y, Tamada T, Roumestand C, Biomol NMR Assign, 2024

An interactive online or local Java tool for homology modelling and protein family study coupling a full featured alignment editor with a complete 3D viewer.
Grand F, Pons JL, Labesse, G (2016)


Several genomes, databases, or protein family have been modeled during various studies with the help of automatic procedure of @TOME. The generated models are accessible from this database.
Reference : Pons JL & Labesse G.(2009) Nucleic Acids Research ; doi : 10.1093/nar/gkp368
This web server is dedicated to the protein structural family of Knottins
Gelly JC, Gracy J, Kaas Q, Le-Nguyen D, Heitz A, Chiche L. (2004) Nucleic Acids Res.;32(1):D156-9.
Postic G, Gracy J, Périn C, Chiche L, Gelly JC. (2018) Nucleic Acids Res.;46(D1):D454-D8.
Additional Sponsors : ACI-IMPBio
CoSiAn (Combinatorial Similarity Analysis)
Similarity analysis between a reference ligand and a bank of ligands to screen, using a combination of 2D and 3D similarity tools.
Victor Reys - Violaine Moreau - Gilles Labesse
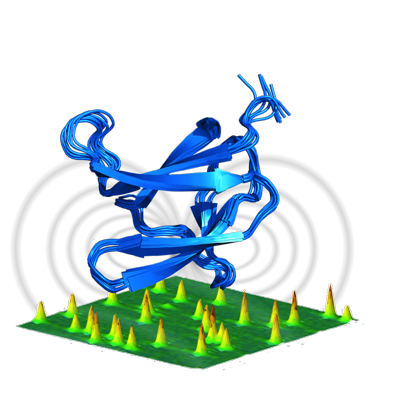
Christian Roumestand and Nathalie Declerck
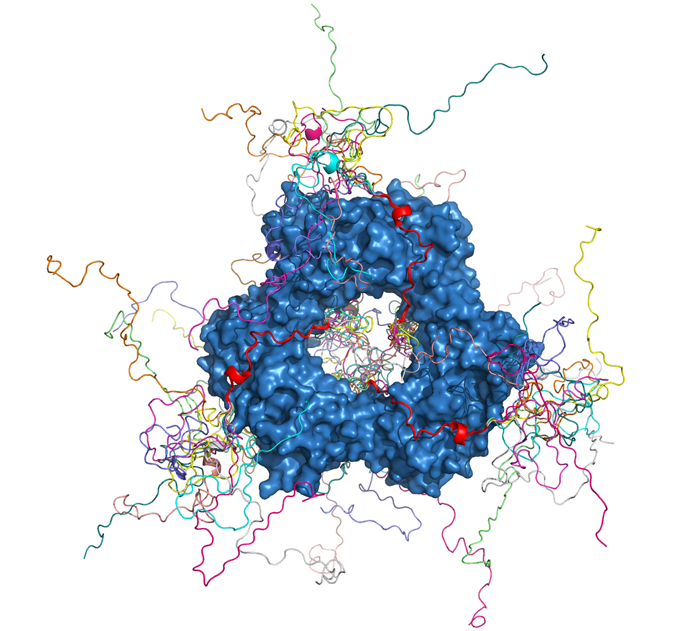
Pau Bernadó and Nathalie Sibille
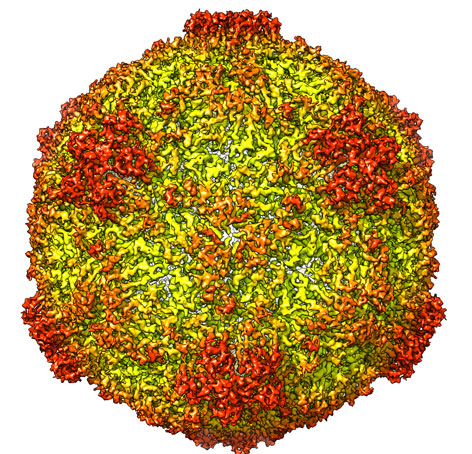
Patrick Bron
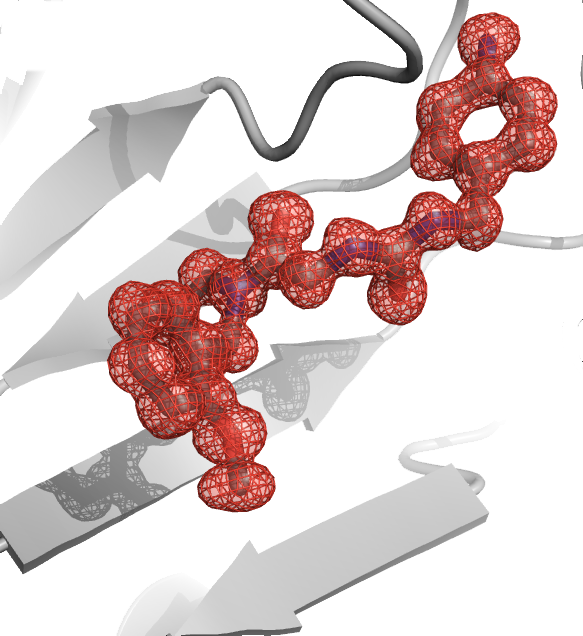
Gilles Labesse
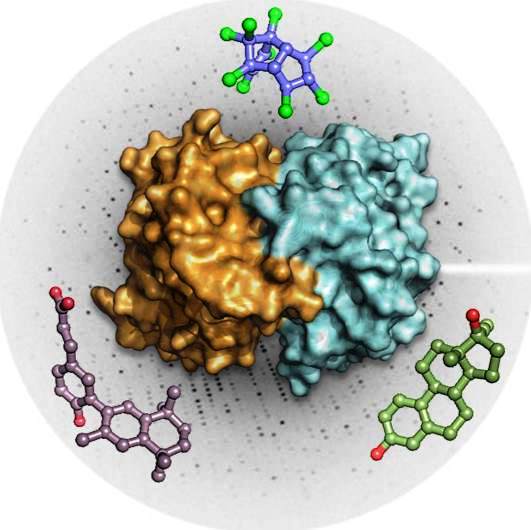
William Bourguet and Albane Le Maire
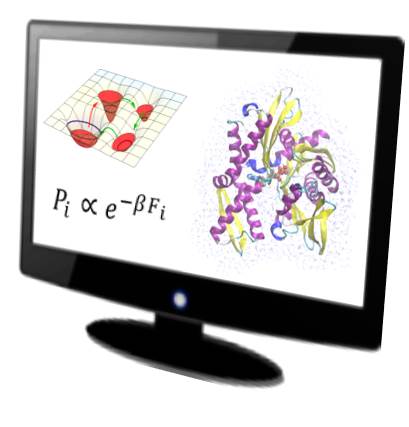
Alessandro Barducci
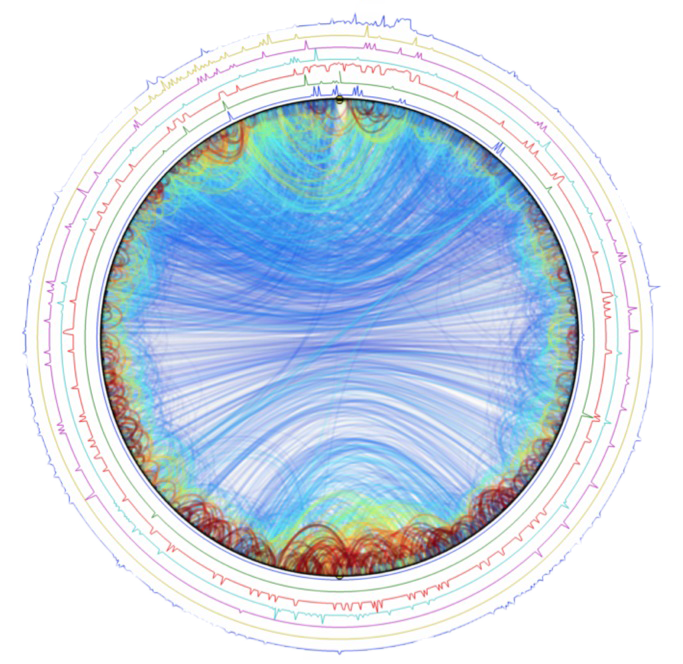
Marcelo Nollmann
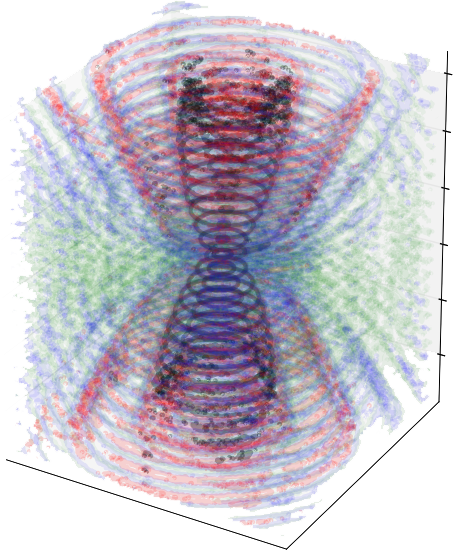
Manouk Abkarian and Francesco Pedaci

Pierre-Emmanuel Milhiet and Emmanuel Margeat
Jérôme Bonnet and Martin Cohen-Gonsaud

Alessandro Barducci
Thomas Robert
Guillaume Cambray
Jakub Gruszczyk