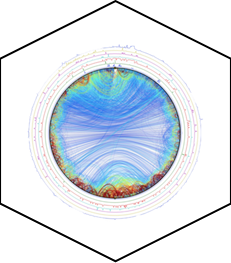
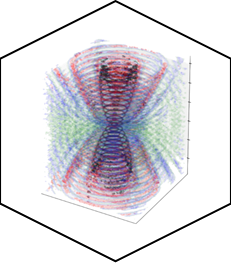
PED in 2024: improving the community deposition of structural ensembles for intrinsically disordered proteins
Ghafouri H, Lazar T, Del Conte A, Tenorio Ku LG, Tompa P, Tosatto SCE, Monzon AM, Aspromonte MC, Bernado P, Chaves-Arquero B, Chemes LB, Clementel D, Cordeiro TN, Elena-Real CA, Feig M, Felli IC, Ferrari C, Forman-Kay JD, Gomes T, Gondelaud F, Gradinaru CC, Ha-Duong T, Head-Gordon T, Heidarsson PO, Janson G, Jeschke G, Leonardi E, Liu ZH, Longhi S, Lund XL, Macias MJ, Martin-Malpartida P, Mercadante D, Mouhand A, Nagy G, Nugnes MV, Perez-Canadillas JM, Pesce G, Pierattelli R, Piovesan D, Quaglia F, Ricard-Blum S, Robustelli P, Sagar A, Salladini E, Senicourt L, Sibille N, Teixeira JMC, Tsangaris TE, Varadi M, Nucleic Acids Res, 2024