Super-resolved 2D and 3D fluorescence imaging
| Service |
Super-resolved 2D and 3D fluorescence imaging
|
|||
| We have two home-made SMLM microscopes to perform STORM and PALM. One is combined with an Atomic Force Microscope and enables correlative fluorescent and topographic imaging at the nanometer scale. | ||||
|
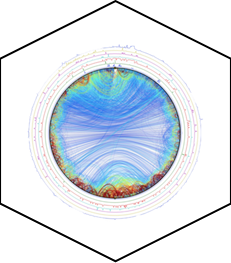
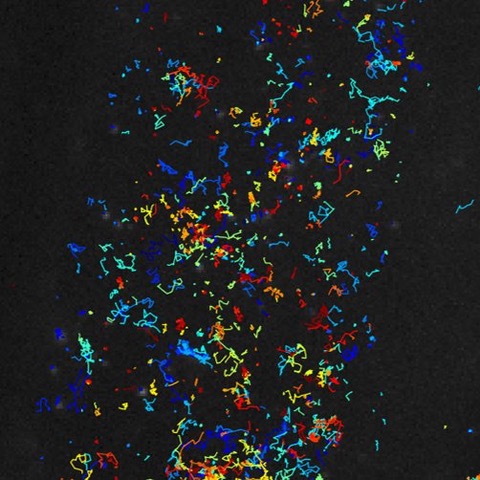
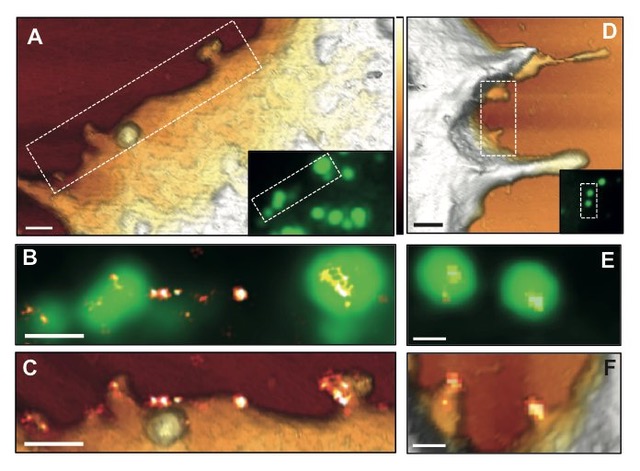
Correlative TIRF STORM AFM image of HIV budding from a HeLa cell - credit: Selma Dahmane |
||||
|
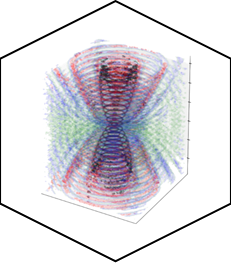
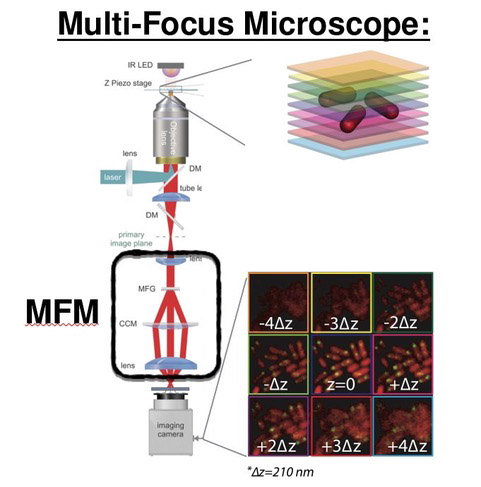
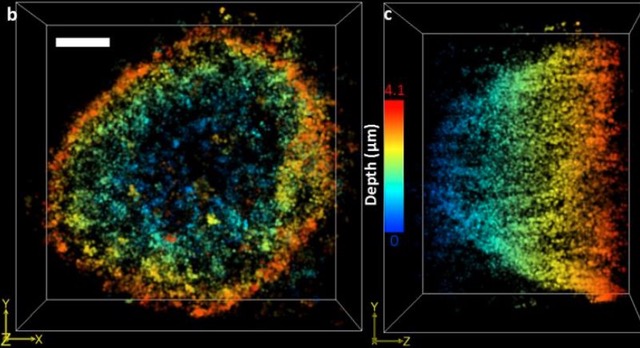
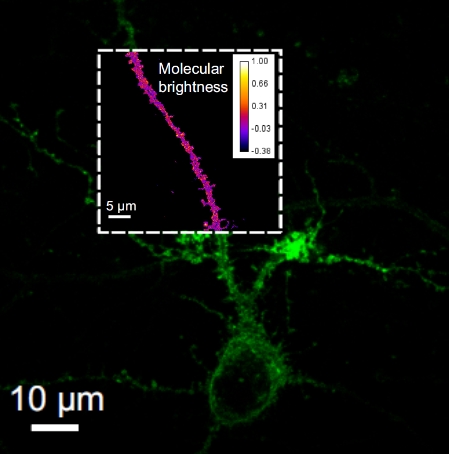
We propose Multi Focal Microscopy (MFM) to produce fast 3D tracking or 3D super-resolved imaging of fluorescently labeled samples. The setup is currently designed for imaging sections of ~3µm (bacteria, yeast) and is still under optimization. |
||||
|
||||
|
A 3D STED setup and a light-sheet microscope are currently under construction on our R&D facility. Stay tuned! All projects are run as collaborations. Please This email address is being protected from spambots. You need JavaScript enabled to view it. for help choosing the most appropriate approach. We also provide advice to design your experiment and support with data analysis. After a meeting, we can run a few experiments to assess the feasibility of the approach. Then a collaboration scheme is defined between the team and the facility. Our R&D division can also implement specific adaptations for your project if needed.
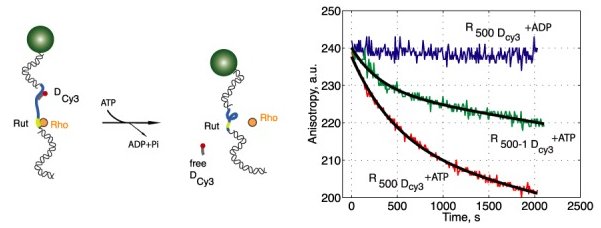
Selected publications: B. Guilhas, J.C. Walter, J. Rech, G. David, N.-O. Walliser, J. Palmeri, C., Mathieu-Demaziere, A. Parmeggiani, J.Y. Bouet, A. Le Gall, M. Nollmann. ATP-driven separation of liquid phase condensates in bacteria, Molecular Cell, 2020 Jul 16;79(2):293-303.e4. pubmed Id 32679076 Selma Dahmane, Christine Doucet, Antoine Le Gall, Célia Chamontin, Patrice Dosset, Florent Murcy, Laurent Fernandez, Desirée Salas Pastene, Eric Rubinstein, Marylène Mougel, Marcelo Nollmann, and Pierre-Emmanuel Milhiet. Nanoscale organization of tetraspanins during HIV-1 budding by correlative dSTORM/AFM. Nanoscale, 2019 Mar 28;11(13):6036-6044 Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M, Bantignies F, Cavalli G. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Science Advances. 2018 Feb 28;4(2):eaar8082. doi: 10.1126/sciadv.aar8082. eCollection 2018 Feb. PMID: 29503869 Salas D*, Le Gall A*, Fiche JB, Valeri A, Ke Y, Bron P, Bellot G, Nollmann M [*co-authors] Angular reconstitution-based 3D reconstructions of nanomolecular structures from superresolution light-microscopy images. PNAS, 2017 Aug 15. pii: 201704908. doi: 10.1073/pnas.1704908114 Sanchez A, Cattoni DI, Walter JC, Rech J, Parmeggiani A, Nollmann M, Bouet JY Stochastic Self-Assembly of ParB Proteins Builds the Bacterial DNA Segregation Apparatus Cell Syst 2015, 1(2):163-73 pubmed Id 27135801 Laura Oudjedi, Jean-Bernard Fiche, Sara Abrahamsson, Laurent Mazenq, Aurélie Lecestre, Pierre-François Calmon, Aline Cerf, and Marcelo Nöllmann. Astigmatic multifocus microscopy enables deep 3D super-resolved imaging. Biomedical Optics Express, 7(6): 2163-2173. Apr 2016 Sara Abrahamsson, Rob Ilic, Jan Wisniewski, Brian Mehl, Liya Yu, Lei Chen, Marcelo Davanco, Laura Oudjedi, Jean-Bernard Fiche, Bassam Hajj, Xin Jin, Joan Pulupa, Christine Cho, Mustafa Mir, Xavier Darzacq, Marcelo Nollmann, Maxime Dahan, Carl Wu, Timothy Lionett, James . Liddle and Cornelia I. Bargmann. Perfect color Multifocus Microscopy (MFM) with increased sensitivity. Biomedical Optics Express, 7 (3): 855-69, 1 March 2016. Vishwakarma, R.K., Cao A.M., Morichaud Z., Perumal A.S., Margeat E., Brodolin K. Single-molecule analysis reveals the mechanism of transcription activation in M. tuberculosis, Science Advances, 2018
|
||||
| Equipement |
PALM
|
TIRF/3D MFM
|
|||||
 |