Optimizing translation efficiency in E. coli
This project leverage genome-scale DNA synthesis to focus on the precise engineering of a small region in reporter constructs.
Our goals are:
Fundamental — to better understand the sequence determinants of a critical cellular process and how they can impact the physiology of the cell.
Applied — to produce a reliable design tool to control and optimize the cost of heterologous production in E. coli.
Both the productivity and stability of artificial genetic circuits depends on the burden that their introduction imposes on the cell.
Protein biosynthesis represents half the energy expenditure of exponentially growing cells. The making of new ribosomes, the molecular machine responsible for the translation of mRNA into proteins, amounts to up to 40% of the total protein synthesis. As a consequence, ribosomes hold great value on the cellular market. They must be allocated wisely over the transcriptome to enable synthesis of other important protein while ensuring the maintenance of a sufficient pool in dividing cells.
The introduction of heterologous genes perturbate the allocation of ribosomes, leading to severe growth defects that extend beyond the direct cost of producing extraneous protein. 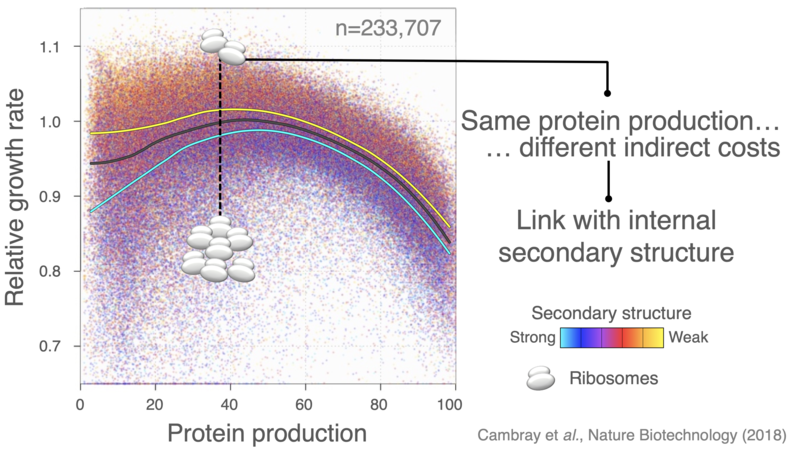
Optimizing translation efficiency to minimize the mobilization of ribosomes provides a general framework to both improve control over heterologous protein yield and minimize indirect production cost, therefore improving genetic stability by limiting the selective advantage of escape mutants that break the imposed expression.
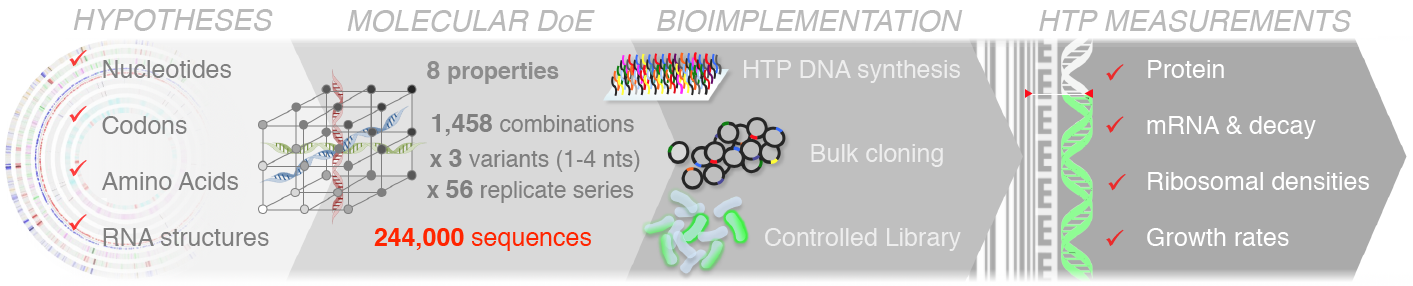
We have used large-scale DNA synthesis to comprehensively explore combinations of sequence properties impacting translation initiation. Construction and characterization of nearly a quarter million sequences has allowed us to disentangle the respective contributions of each of these properties on the synthesis of a reporter, but also on the stability of its mRNA, its propensity to sequester ribosomes and consequent physiological impacts on cell growth. Our results, have put forward important feedback loops between the amount of protein produced, the stability of messenger RNAs and the cost of production.
We are now extending this high-throughput approach to the identification of coding sequence properties affecting translation elongation, with particular attention to the consequences on messenger RNAs stability.
We are combining machine learning and mechanistic modeling to obtain a global understanding of the translation process through our data. Our eventual goal is to develop a data-driven gene sequence optimization tool capable of predicting expression levels and maximize the protein production/physiological cost ratio.
Collaborators: Luca Ciandrini (CBS), Sebastien Nouaille and Laurence Gribal (TBI)
2-years position post-doctoral position available (need link)