Photo-Activable Localization Microscopy (PALM/ STORM)
Associated to the group of Marcelo Nollmann
The image that a point source makes on a camera is called the point-spread function (PSF). The PSF is limited by diffraction to be no less than approximately half the wavelength of light. Until very recently, the spread of the PSF defined the maximum resolution attainable by conventional fluorescence microscopy.
PALM and dSTORM are imaging techniques achieving resolution well below the diffraction limit, the 2D resolution being usually between 30 and 50nm (see Betzig et al. 2006 – Hess et al. 2006 – Gould et al. 2009). These techniques rely on the fact the position of single fluorescent emitters can be calculated very precisely by measuring the shape of their PSF, the localization error being inversely proportional to the square-root of the number of photons collected. Super-resolution is achieved using fluorescent labels that are stochastically activated during the measurement. Therefore, at a given time, only a small subset of dyes is emitting, the majority remaining in a “dark” state. Single fluorescent events are detected and their mean positions calculated. After repeating this activation/emission/bleaching cycle many times, a reconstructed high-resolution image of the sample can be calculated using the position of all the molecules detected during the measurement.
PALM usually refers to super-resolution microscopy using photo-activable fluorescent proteins (PAFP) such as mEos2 or Dronpa (see Lippincott-Schwartz et al. 2009 - Fernández-Suárez et al. 2009). PALM is particularly well suited for in vivo imaging. However, the preparation of a fusion between the protein of interest and a PAFP is required.
On the other hand, dSTORM uses synthetic dyes such as Cy5 or Atto647 (Heilemann et al. 2009 – van de Linde et al. 2011). In that case, the activation relies on the formation of a complex between the dye and thiolated compounds, sending the fluorophore into a dark state. These techniques are very well suited for in-vitro experiments or with fixed cells. Immunostaining and specific switching buffers are necessary for this technique.
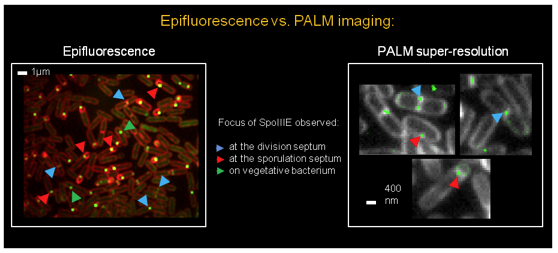
Applications and developments
Our PALM microscope allows for the investigation of assembly and dynamical movement of fluorescently labeled protein complexes in fixed or live cells such as Bacillus subtilis or Drosophila melanogaster.
Inverted microscope Zeiss with a 100x oil-immersion objective (NA = 1.45). Four wavelengths of excitation and activation are available: 641nm (100mW), 561nm (100mW), 488nm (50mW) and 405nm (50mW). Both Epifluorescence and TIRF illuminations are possible on the setup. The focus is controlled in real time by Back Focal Plan Reflection of a 785nm IR laser.
Below is a list of the filters available on the setup:
New developments are in course to actively stabilize our PALM microscop, adapt it for 3D-PALM, and concentrate the excitation light onto the area of interest to reduce background excitation and increase the power density of the excitation beam. To this aim, we will control the shape and three-dimensional position of the excitation beam in the sample plane by using a spatial light modulator (SLM), a computer controlled optical device that allows for the phase and intensity modulation of the light beam. This device allows for the formation of arbitrary shapes in or out of the focal plane and will permit us to excite an arbitrary region of the cell body and thus considerably reduce background fluorescence.
Our setup
New Developments

