Pau Bernado
Collaboration : Stéphane Delbecq (LBCM, Montpellier) and Bente Vestegaard (U. Copenhagen)
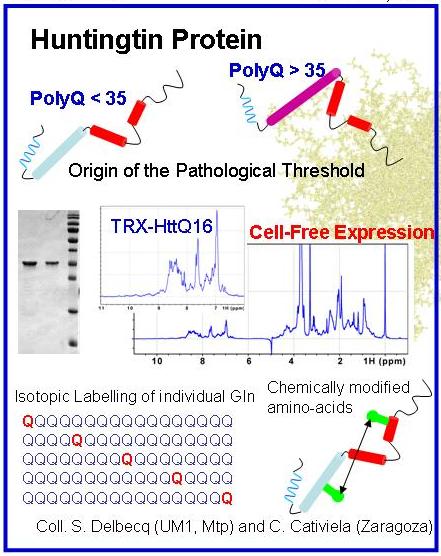
Huntington’s disease (HD) is a rare neurodegenerative pathology caused by mutation of the huntingtin gene that results in a protein with an expanded stretch of glutamines (>35) with a high propensity to form amyloidogenic fibres. Our aim is the structural characterization of the relevant species of huntingtin involved in HD. This project is funded by the ANR (SPIN-HD, CHEX 2011).
The N-terminal region huntingtin is an IDP in its native state. Its structural characterization will be performed by combining state-of-the-art NMR and SAXS experiments in pathological and non-pathological protein variants. The structural models of huntingtin derived from the integration of all experimental data will unravel the molecular origin of the instability in proteins with expanded glutamine tracts. In collaboration of Juan Cortés (LAAS-Toulouse), the use of robotics-inspired algorithms will be tested to study the structure and conformational dynamics of polyGln tracts.
The oligomeric species found at the initial stages of the aggregation of huntingtin are hypothesized to be the toxic agents in HD as shown in other neurodegenerative diseases. The structure and the origin of the toxicity of these species remain elusive due to the dynamic nature of amyloidogenesis. Mathematical analysis of SAXS measurements performed during fibril formation will allow the deconvolution of the data solving the intrinsic polydisperisity problem. From this analysis, the structural details of the species involved in fibrillation and the kinetics of the pathway will emerge. The effects on the aggregation of small molecules known to delay or stop fibrillation will be monitored using this approach. Strategies designed to pursue this project will be of general applicability, and as a long-term objective they will be exported to other polyGln diseases and other neuropathologies such as Alzheimer and Parkinson.
Nathalie Sibille
Collaboration : Bernard Mouillac, Team "Récepteurs membranaires : structure, dynamique et pathologie" Institut de Génomique Structurale (IGF), Montpellier
G Protein-Coupled Receptors (GPCRs), also known as seven-transmembrane α-helix (7TM) domain receptors, are the largest family of membrane proteins found in mammalian cells. They sense a large variety of stimuli such as hormones, nucleotides, lipids, ions, photons and neurotransmitters outside the cell, and activate cellular responses via various signal transduction pathways. GPCRs are involved in numerous human pathologies, and are thus the target of approximately 40% of all current pharmaceuticals. To better design drugs that target GPCRs, it is crucial to fully understand how these receptors function.
An important step toward understanding GPCR function came with the tridimensional structure determination of the Beta-2 adrenergic receptor (β2AR). This structure came out seven years after the structure of Rhodopsin, the first membrane protein solved by X-ray crystallography. The β2AR structure was followed by crystal structures of a number of other GPCRs.
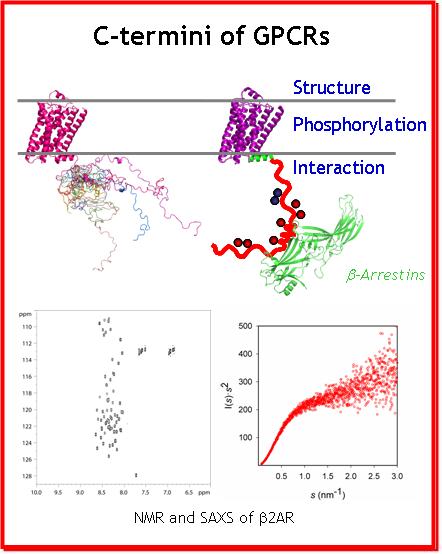
Beside a common core of seven transmembrane helices (7TM), the GPCRs harbor N- and C-termini, that could not be crystallized due to their intrinsic flexibility. Hence little structural data is available for these regions of GPCRs which are, however, crucial for proper signal transduction: desensitization, internalization (endocytosis) and other signaling (scaffolding) functions. These functions are mediated by the interaction of the C-ters with β-arrestins, implicated in important signalling pathways, independent of those related to G proteins which were long thought as the only cytoplasmic partners of GPCRs. In addition, the functional interactions of the GPCR C-ters with β-arrestins are modulated by the phosphorylation of the C-ters by the G protein coupled Receptor Kinases (GRKs). In addition, the behaviour of the intracellular GPCR C-ters is regulated by the extracellular binding of ligands to the surface. Accordingly, C-ters appear as a hub for subtle regulation and signal transduction in a poorly understood manner.
It is noteworthy that the role of GPCR C-ters and the importance of IDPs, became of high interest in cell biology during the last decade. This project will focus on the structural characterization and comparison of four GPCR's C-termini, in their phosphorylated and unphosphorylated forms, by NMR. This study will reveal the key structural and dynamic features of GPCR C-ters, as well as the impact of phosphorylation on these properties that modulate the recruitment of β-arrestins. This information will be key to understand and rationally modulate cellular signal transduction of GPCRs in the future.
Other Projects
Pau Bernadó : Advanced Computational Approaches to study protein motions.
Collaboration : Juan Cortés (LAAS – Toulouse)
The study of flexible systems at atomic level requires detailed description of the structure and the mechanisms inducing their fluctuations. In that context, our group is interested in advanced computational approaches allowing the exploration of the conformational space in an efficient manner to study mobility in broad time-scales. We are interested in Normal Mode approaches that perturb 3D structures in a physically meaningful manner. Derived conformations can be evaluated using SAXS data, which are exquisitely sensitive to large conformational fluctuations.
Robotics-inspired algorithms have been demonstrated to be very efficient in sampling the 3D space available to macromolecules. In collaboration with Juan Cortés (LAAS-Toulouse) we aim to apply these approaches to study conformational transitions in IDPs.
Nathalie Sibille : RDC under pressure
Collaboration : Christian Roumestand and Cathy Royer, CBS, Montpellier
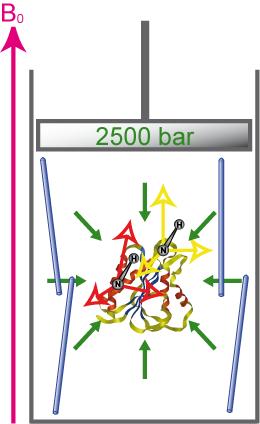
Accurate description of protein folding and stability, in normal and pathological conditions, has been a major area of biophysical chemistry for over 50 years. Although much progress has been made in recent years to achieve this objective, a number of important questions remain. The physical basis of pressure effects on the structure and stability of the protein remained a mystery unlike the effects of temperature.
We have recently characterized several alignment media that are amenable to high-pressure. This enables the measurement of RDCs in a broad range of pressures that will enable the structural characterization of protein states emerging under these conditions. For more details see :
Sibille N*, Delarole M, Royer C, Roumestand C. Measuring Residual Dipolar Couplings at High Hydrostatic Pressure : Robustness of Alignment Media to High Pressure. J Biomol NMR. (2014).

