Our projects aim at bringing a quantitative and integrative view on molecular mechanisms of transcription regulation in bacteria, combining in vivo and in vitro approaches.
Prokaryotic transcription termination
N. Declerck, E. Margeat, C. Clerté, R. Vishwakarma
This work focuses on the two mechanisms used by bacteria to terminate transcription of a gene (intrinsic termination and Rho-dependent termination.
We use a combination of single molecule observation (smFRET, SPT-PALM & correlation spectroscopies), nano-manipulation and structural biology to study the dynamics events that lead to transcription termination in bacteria. Specifically, our research focuses on:
1. LicT/SacY antitermination proteins from Bacillus subtilis, that prevent the premature arrest of transcription by binding to an antiterminator RNA hairpin that overlaps an intrinsic terminator.
2. The transcription terminator Rho, an ATP-fueled hexameric helicase and translocase, implicated in the dissociation of transcription elongation complexes.
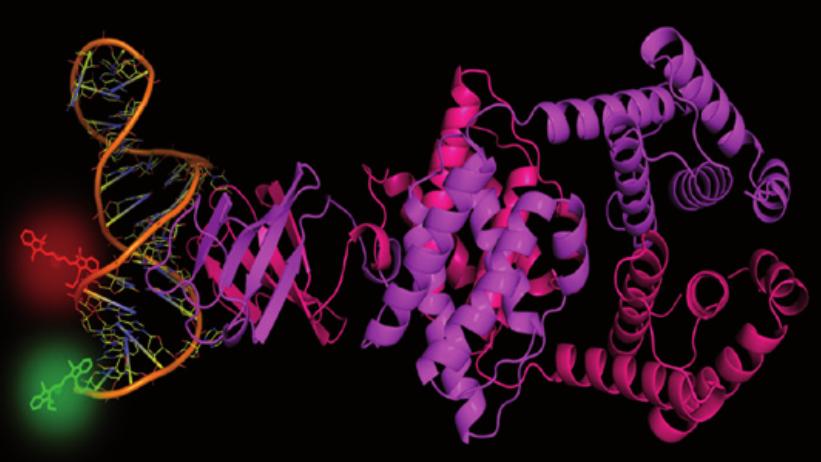
Cartoon representation of the LicT antitermination protein dimer, bound to its target RNA hairpin that has been labeled with a pair of fluorophores to monitor its conformational dynamics in a single pair FRET experiment.
Collaborations: H. Demene, F. Pedaci, M. Nollmann (CBS), M. Boudvillain (CBM, Orleans), N. Figueroa-Bossi (I2BC, Gif/Yvette), K. Brodolin (IRIM, Montpellier)
Bacterial adaptation to environmental changes
N. Declerck, A. Bourges, C. Clerté
Our aim is to unravel the molecular mechanisms that underlie the adaptation of bacteria to rapid changes in their environment, from the atomic and single molecule resolution to the level of single cells and bacterial populations.
We particularly focus on the control of glycolysis/gluconeogenesis by the CggR and CcpN repressors from B. subtilis. We use highly sensitive and quantitative fluorescence fluctuation-based microscopy methods to measure directly the intracellular concentration, oligomerization state and diffusion properties of these regulatory proteins in live bacterial cells. Combined with in vitro experimental and modelling approaches, our studies provide an integrated picture of the adaptive response to a nutritional shift. We apply similar approaches for characterizing the Mrr endonuclease responsible for the activation of the SOS response in E. coli cells exposed to high pressure.
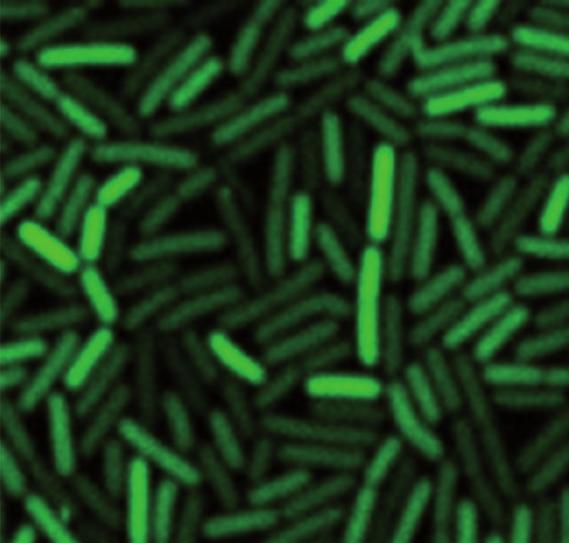
Live cell imaging of a B. subtilis strain expressing a PccgR-GFP transcriptional fusion under repression on malate.
Collaborations: S. Aymerich (INRA, Jouy-en-Josas), C. Royer (RPI, NY), O. Radulescu (UM, Montpellier)
G. Bellot, E. Margeat, N. Aissaoui, A. Mills
We use DNA self-assembly methods that offer 3D-shape control at nanoscale resolution and programmable actuation. Our aim is to develop new bio-inspired intelligent systems to help solve problems of fundamental and medical interest.
DNA nanotechnology. DNA origami represents a major landmark as the first method to self-assemble megadalton scale nanostructures with arbitrarily-defined morphology. Our main objective is to explore new design principles for self-assembling molecular architectures for de novo fabrication of high-performing materials.
Membrane proteins. The understanding of the molecular mechanisms of the cell membrane requires new experimental tools at the nanometre level. We aim to develop DNA nanostructures to facilitate structural studies of membrane proteins by cryo-EM and single-molecule fluorescence microscopy.
Bio-inspired nanomechanics. Inspired by the biomechanical features of protein assemblies, our goal is to build nanometer-sized structures that exhibit controllable motions and functions. These man-made nanodevices can be used as sensors that respond to specific stimuli and may find applications in areas such as biosensing, biophysics and on-demand drug delivery.
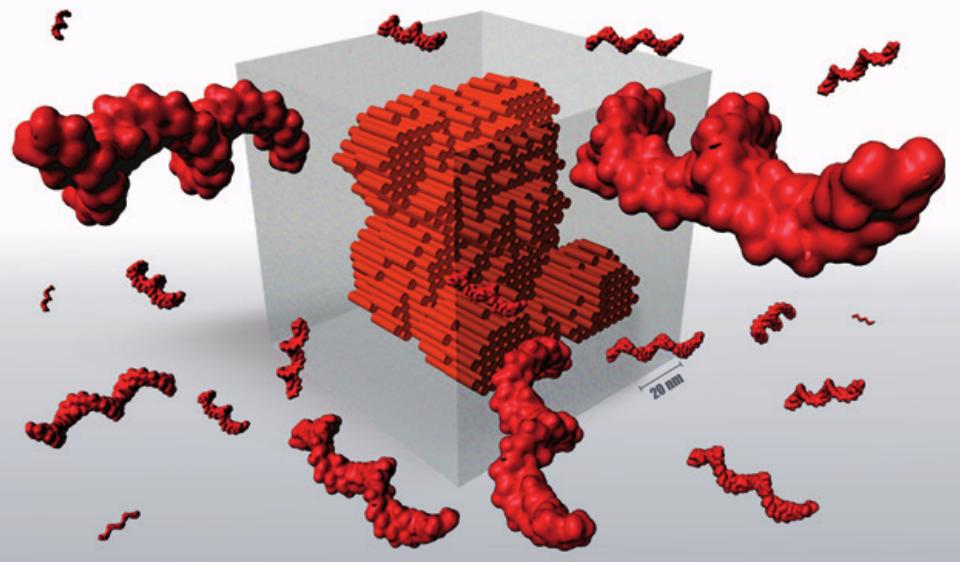
DNA self-assembly of 3D nanostructures from 1.5 million of nucleobases.
Collaborations: Y. Ke, (Georgia Tech, USA), S. Granier (IGF, Montpellier), S. Bidault, (ESPCI, Paris).

