C. Lionne, M. Gelin, G. Labesse
The goal of the group is to help the design of efficient and specific drugs by studying enzyme reaction pathways and drug-target interactions by the mean of fine enzymology, biophysical characterization and thermodynamic approaches.
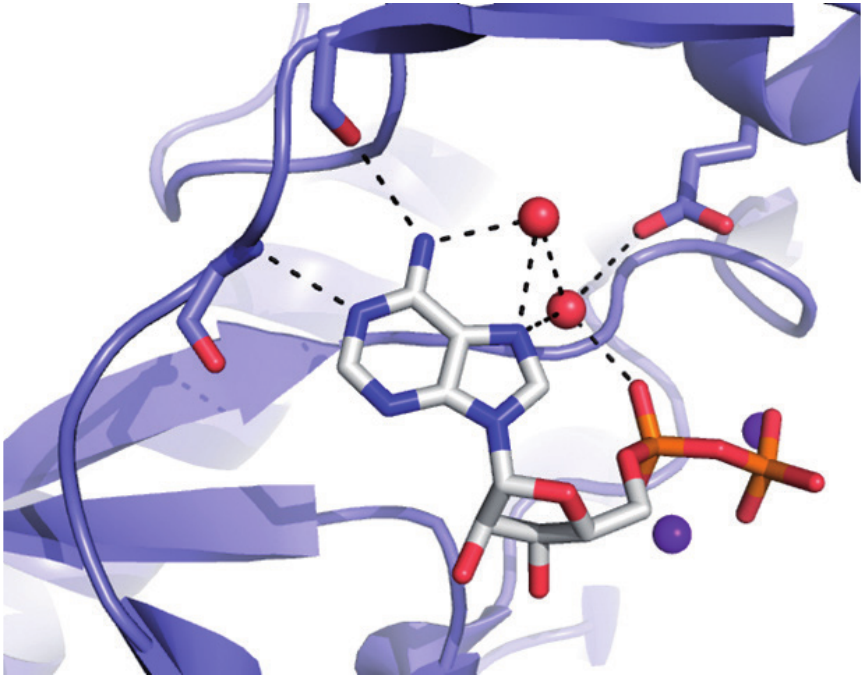
Bacterial antibiotic modifying enzyme: identification of the main reaction intermediate to be targeted.
Efficient and specific drugs directed against enzymes are much easier to design with a deep understanding of how the proteins work, and especially in which states they spend most of their time. Transient kinetics techniques (stopped-flow, quench-flow, cryoenzymology) are routinely used to measure protein-ligand kinetics (kon, koff ), identify reaction pathway and rate-limiting steps, as well as to characterize the effects of inhibitors thereon. Beside, other biophysical techniques are used to characterize drug-target interactions: classical steady state enzymology, isothermal titration calorimetry, thermal shift assay, molecular docking and X-ray crystallography. Optimized inhibitors are designed and their biological effects tested (in collaboration for cellular and animal models). The group focuses its research on new targets working on nucleos/tides for the design of innovative anti-infectious or anti-cancer therapies.
Collaborations: Zohra Befodda (CHROME Nîmes), Anne Blanc-Potard, Rachel Cerdan & Sharon Wein-Gratraud (LPHI Montpellier), Konstantin Brodolin & Laurent ), (VBIC Nîmes), Jean-Michel Jault et Elise Kaplan (IBCP Lyon), Thierry Lorca (CRBM Montpellier), Suzanne Peyrottes & Christophe Mathé (IBMM Montpellier), Sylvie Pochet & Olivier Dussurget (Institut Pasteur Paris), Katharina Rox (HZI Braunschweig).
Recent references: Praharaj et al. Sci Adv. in press; Kaplan et al. ChemMedChem. 2025; EMBO J. 2024; Nat Commun. 2023; Clément et al. Eur J Med Chem. 2023; Rahimova et al. FEBS J. 2023; Mary et al. Mol Cell. 2022.

