Theme 3: Nuclear receptors as targets of environmental endocrine disruptors
C Carivenc, W Bourguet
Many chemicals released into the environment bind to NRs and activate or inhibit their action in an inappropriate spatio-temporal fashion. Referred to as endocrine disrupting chemicals (EDCs), these compounds cause a wide range of developmental, reproductive, neurological or metabolic defects. The goal of this project is to provide mechanistic insights into the endocrine disrupting action of environmental pollutants.
The group of molecules acting as EDCs is highly heterogeneous and comprises compounds that are often distantly related to endogenous ligands. It contains substances as chemically different as bisphenols, phthalates, parabens, dioxins, alkylphenols, organotins, etc. This large structural diversity renders the interaction of EDCs with NRs poorly understood and barely predictable. Our correlative analysis of structural, biophysical, cell-based and in vivo data allows revealing a variety of, sometimes unforeseen, mechanisms of action. Launched in 2009, this project is now progressing with the elucidation of the structures of many EDCs in complex with various NRs. It is intended to provide a wealth of tools and information for the development of safer chemicals devoid of NR-mediated activity and for environmental risk assessment through the development of robust in silico screening methods.
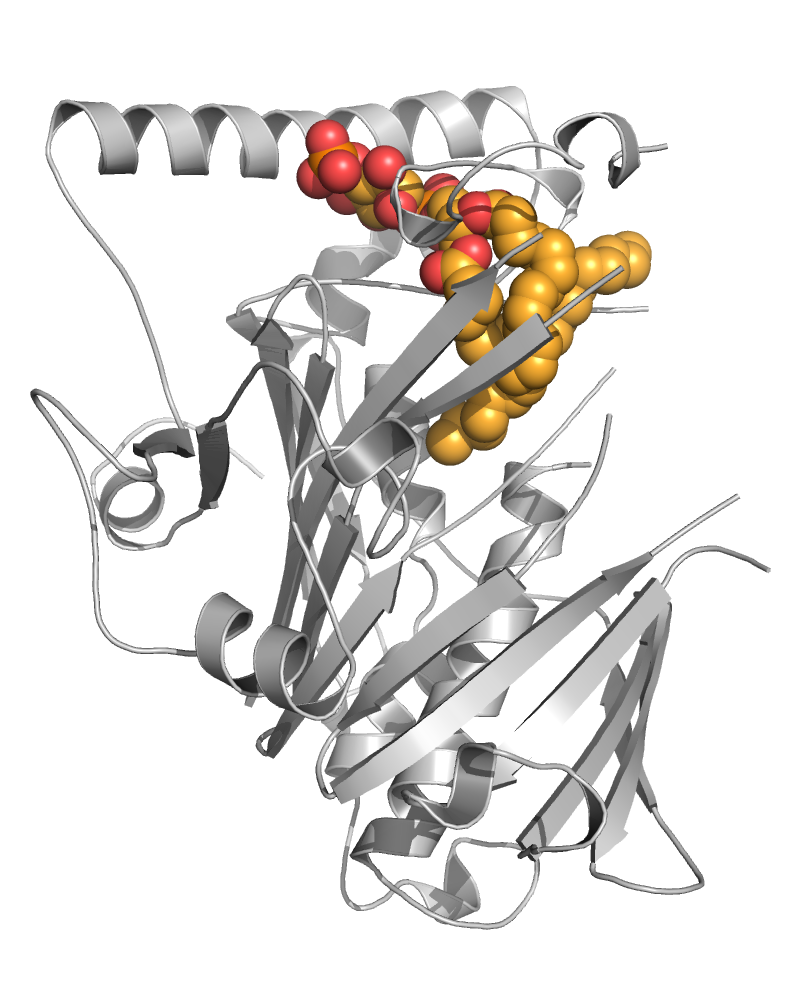
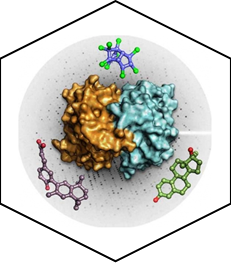
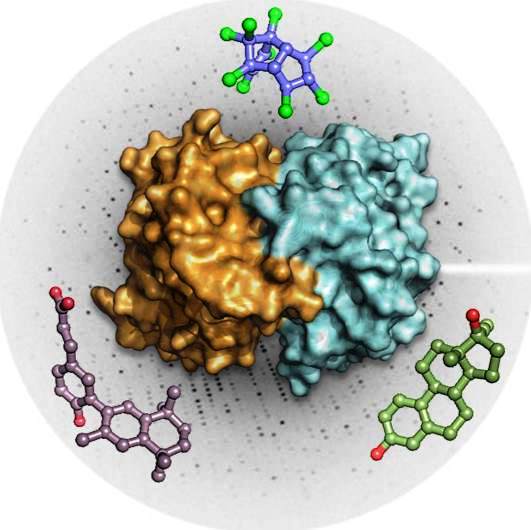
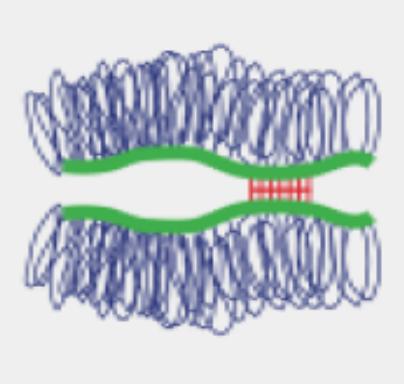
The team has provided mechanistic insights into the interaction of EDCs with several of their major targets including the estrogen receptors (ERα and β), the estrogen-related receptor (ERRγ) and PXR. More specifically, we have reported high-resolution crystal structures revealing the various mechanisms by which distantly related chemicals bind to ERs and act in a subtype-dependent fashion as full agonists, partial agonists or antagonists [Delfosse, PNAS, 2012 ; Delfosse, Environ Health Perspect, 2014]. Our work on ERRγ allowed us to describe an unusual mechanism by which compounds such as bisphenol-A (BPA) bind to this constitutively active receptor which is further activated in an activation helix (H12)-independent fashion. In this particular case, ligand binding induces a global stabilization of the protein, including functional loop regions [Thouennon, Cell Mol Life Sci, 2019]. However, the most exciting achievement of this project has been the discovery of a mechanism for the so-called "cocktail effect" by which chemicals can combine to form dangerous cocktails from harmless components [Delfosse, Nat Commun, 2015]. We have been able to show that two compounds, poorly active on their own, can bind cooperatively and activate synergistically the receptor PXR so that significant activation can be observed at concentrations two orders of magnitude below those required for individual compounds. As an extension of this pioneering work, we have described previously unreported binding mechanisms, substantiating the versatility of PXR's ligand binding domain and the receptor's role in sensing a diverse array of chemicals. In addition, we have shown that environmental compounds targeting RXR, the obligatory heterodimerization partner of PXR, can further boost the activity of two synergizing PXR ligands. These findings support the notion that certain chemicals such as endocrine disruptors or pharmaceuticals, at levels deemed individually safe, can potentially interact synergistically and disrupt critical cell signaling [Delfosse, PNAS, 2020].
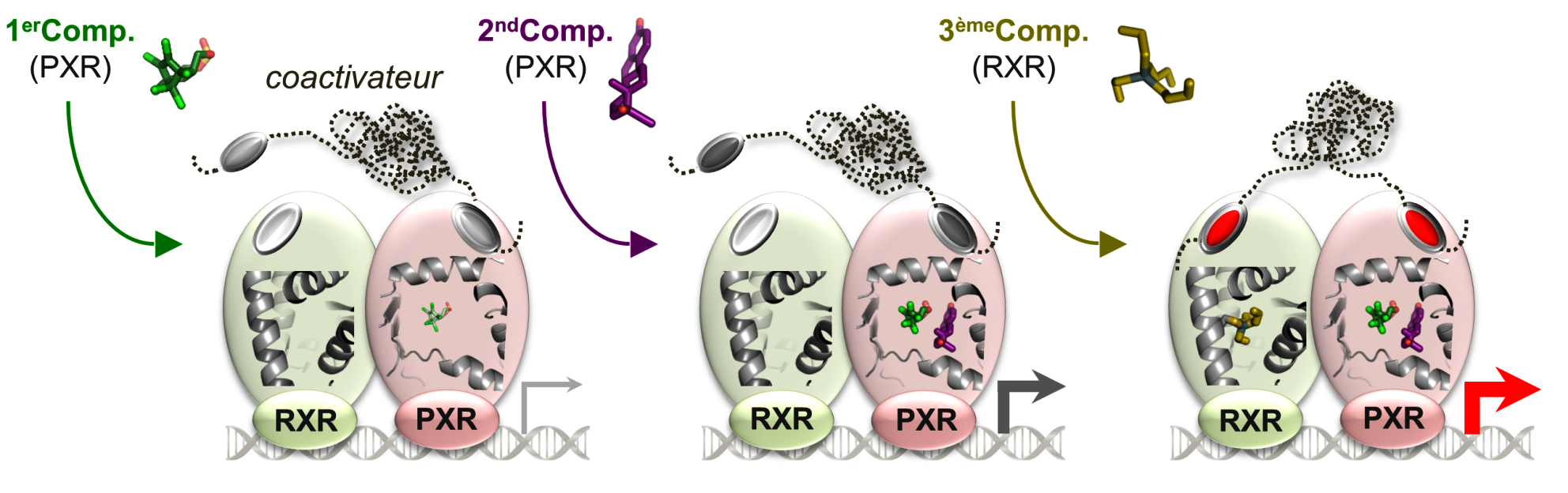
PXR can be activated synergistically by exogenous compounds via two converging cooperative interaction mechanisms involving (1) the binding of two mutually stabilizing compounds in the PXR pocket (center) and (2) the cooperative binding of coactivators through the creation of a second anchor point on the heterodimer when fixing a third substance, on RXR (right).
Beyond demonstrating molecular mechanisms of regulation, we were also engaged in several studies aimed at generating tools for assessing the endocrine disrupting potential of chemicals either experimentally [Grimaldi, Front Neurosci, 2015 ; Grimaldi, Front Endocrinol, 2015] or using computer-assisted methods. The server developed with Team 4 called EDMon (Endocrine Disruptor Monitoring) is already available (http://edmon.cbs.cnrs.fr/). To date, it allows screening for ERα, ERβ and PPARγ and predicts affinities using docking and scoring approaches based on machine learning [Schneider, Bioinformatics, 2020 ; Schneider, Endocrinology, 2019].
Main collaborators: P Balaguer (IRCM Montpellier), B Demeneix (MNHN Paris), V Laudet (OOB Banyuls/Mer), Teams 4 and 6 (CBS)