This project is at the crossroad between synthetic genomics and deep mutational scanning.
Our goals are :
Fundamental — to systematically profile functional and adaptive landscapes at the genome scale and illuminate the tension between robustness and evolvability for entire organisms.
Applied — to explore sustainable alternatives to chemical insecticides based on the reprograming of potential biocontrol agents. This could improve food quality and security, reduce the incidence of major diseases and nuisances, while limiting untargeted environmental impacts and mitigating resistances.
Parvoviruses are amongst the smallest known viruses, and provide excellent systems to measure the functional and evolutionary consequences of mutations at the genome scale. Their single-stranded DNA genome contains a handful of genes involved in replication and capsid formation. It is packaged in non-enveloped capsids that are directly involved in genome protection and targeted delivery. These properties have driven the bioengineering of AAV parvoviruses into one of the most promising gene therapy system.
Densoviruses, a large subfamily of parvoviruses, are associated with non-vertebrate hosts. Opportunities of leveraging this diversity for human health and wellness have largely been untapped.
We are focusing on two densoviruses that represent extreme genome sizes and complexities:
AalDV (4 kb) is pathogenic for the yellow fever and tiger mosquitoes, both major disease vectors (Aedes ægypti and Aedes albopictus).
JcDV (6 kb) is pathogenic for the fall armyworm (Spodopetra Frugiperda), an invasive lepidopteran crop pest.
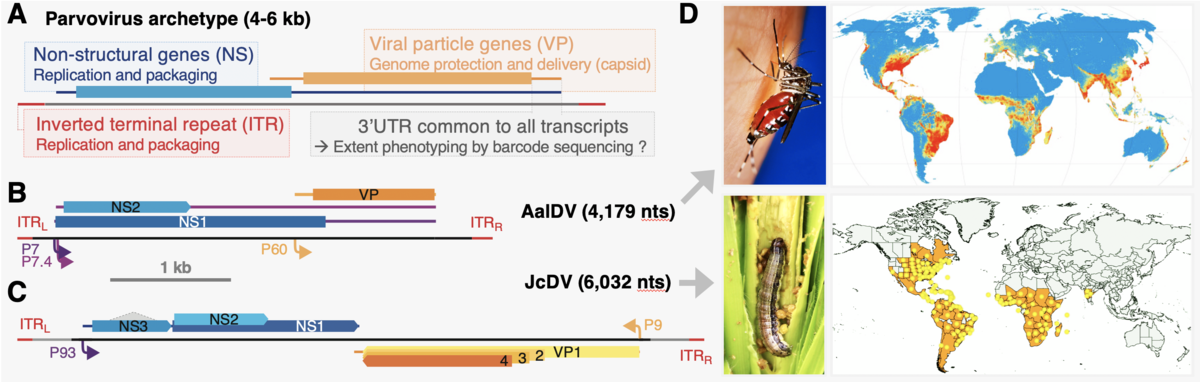
- Models organisms and societal relevance. Parvoviruses display ideal features for genome-wide mutational scanning (A). Genomic organization and expression strategy of Aedes albbipctus densovirus (AalDV, B) and Junonia Coenia densovirus (JcDV, C). Lines represent transcripts; rectangles: coding sequences; colored arrows: promoters; dotted lines: splicing. Features are depicted at scale (see 1kb bar). Both viruses could be used for biocontrol of invasive insect pests (D).
Such viruses hold great potential for controlling populations of nefarious insects. Their smoother and larger capsids could also provide less immunogenic vehicles, with larger packaging capacities, for human gene therapies. Although small, densoviruses are subject to the mechanistic complexities of integrated replicative systems that must interact with their cognate host organisms. Refined molecular understanding is required to firmly establish safety and control for such applications.
We are using large pools of synthetic oligonucleotides to construct libraries that comprise every possible substitutions, insertions and deletion at the nucleotide level. Mutants are tagged with unique DNA barcodes to enable massively parallel phenotypic assays, based on bulk screening and deep sequencing.
Using this high-throughput approach, we can quantify the consequence of mutations on gene expression, RNA stability, splicing, replication, packaging, capsid stability, cell entry, nuclear delivery as well as more integrated phenotypes such as virulence, pathogenicity and host specificity.
Collaborators: Mylène Ogliastro and Anne-Sophie Gosselin (DGIMI); Fabrice Chandre (MIVEGEC).
2-years position available (post-doc or research engineer)

