• Structure and interactions of bacterial antiterminators •
Coordinators: Hélène Déméné
The Bacillus substilis LicT antiterminator protein represents the prototype in gram+ bacteries of widespread regulators of the BglG antitermination familiy. LicT regulates expression of the bglS gene and bglPH operons involved in the transport and metabolim of b glucosides. When activated, these transcriptional regulators bind to a ribonucleotidic antiterminator (RAT) sequence in targets mRNAs, preventing the formation of an antitermination hairpin associated with premature arrest of transcription. All members of the LicT family present a modular structure composed of a RNA binding domain (CAT) and two regulatory homologous domains PRD1 and PRD2. These two domains are the subject of concurrent phosphorylations by enzymes of the PTS (phosphoenolpyruvate PhosphoTransferase System) that control the activation (RNA binding capacity) of LicT. We have previously established the conditions of stability of truncated proteins (CAT-PRD1) and proposed a model of activation for these proteins. We now investigate on the structure of the whole protein, either alone or in interaction with its PTS partners. Our collaborators focus on the structural mechanism at the RNA level.
| Figure : models of activated (closed ) and inactive (open) LicT-CAT-PRD1. The degree of opening of the RNA binding domain determines the level of RNA binding activity. | 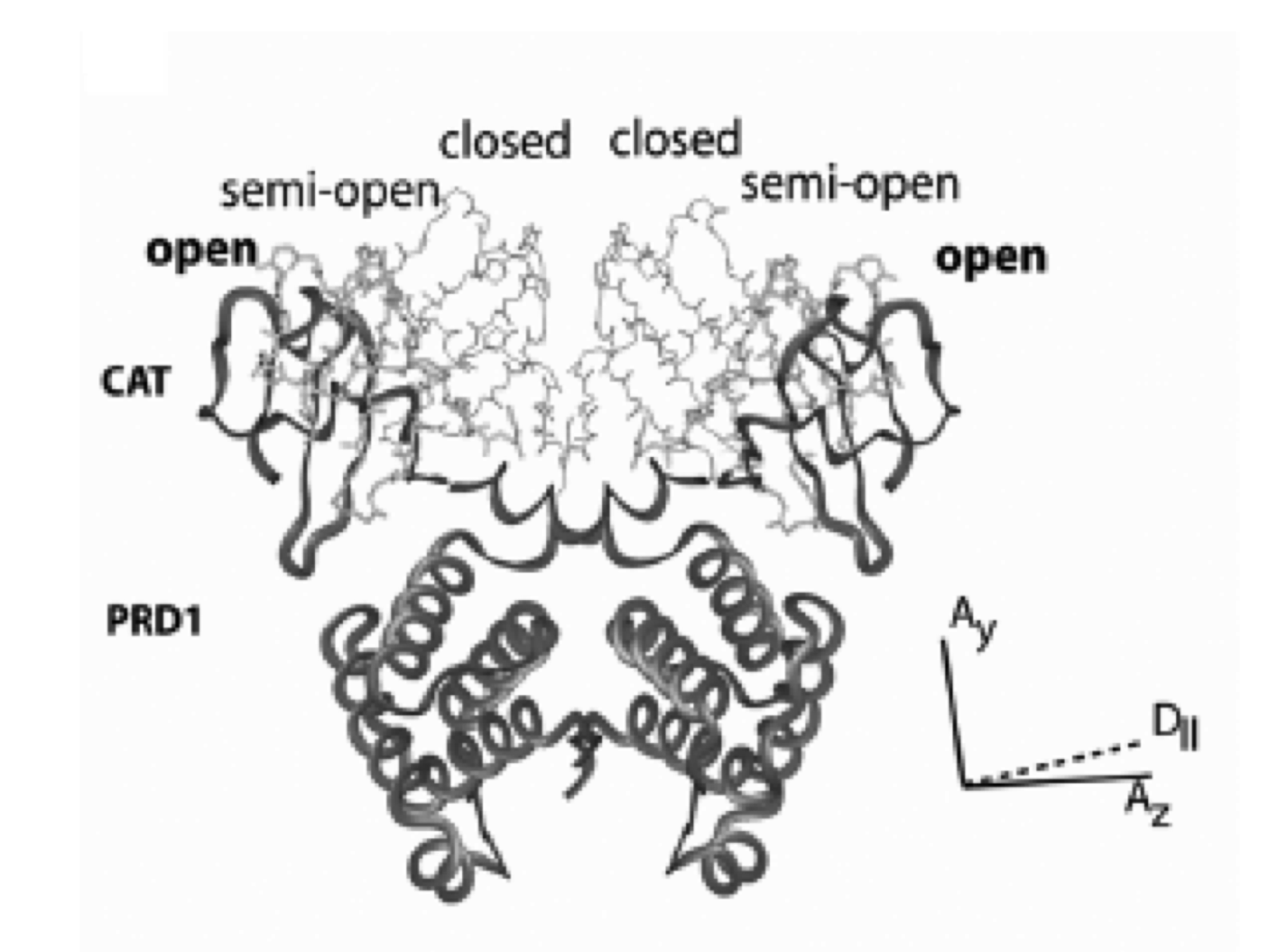 |
NMR Staff : Yang Yinshan
Coll.: Nathalie Declerck (CBS)

