We are developing tools to program living cellular behavior and applying them to healthcare problems. Bellow is a summary of our recent research.
Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates.
Alexis Courbet, Drew Endy, Eric Renard, Franck Molina, and Jérôme Bonnet, Science Translational Medicine, 2015 May 27;7(289):289ra83.

Whole-cell biosensors have several advantages for the detection of biological substances and have proven to be useful analytical tools. However, several hurdles have limited whole-cell biosensor application in the clinic, primarily their unreliable operation in complex media and low signal-to-noise ratio. We report that bacterial biosensors with genetically encoded digital amplifying genetic switches can detect clinically relevant biomarkers in human urine and serum. These bactosensors perform signal digitization and amplification, multiplexed signal processing with the use of Boolean logic gates, and data storage. In addition, we provide a framework with which to quantify whole-cell biosensor robustness in clinical samples together with a method for easily reprogramming the sensor module for distinct medical detection agendas. Last, we demonstrate that bactosensors can be used to detect pathological glycosuria in urine from diabetic patients. These next-generation whole-cell biosensors with improved computing and amplification capacity could meet clinical requirements and should enable new approaches for medical diagnosis.
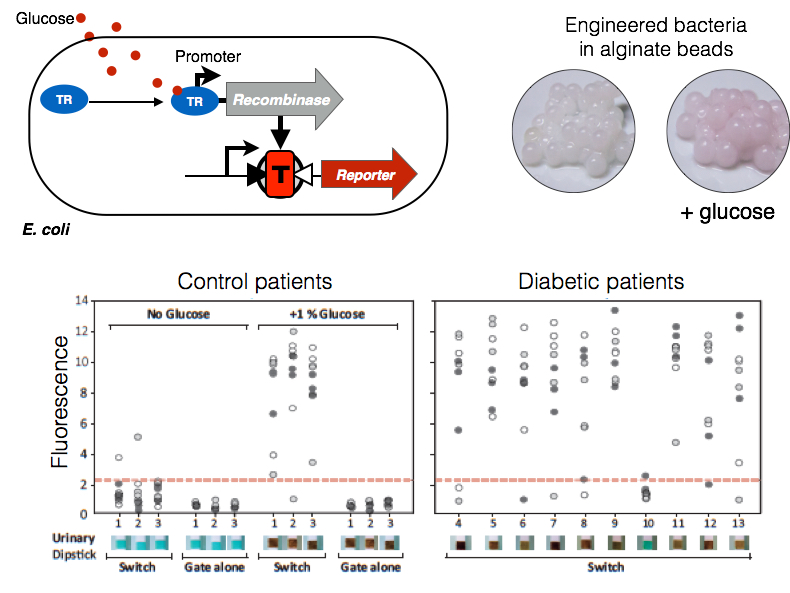
Architecture and operation of a bactosensor detecting glucose. Upper panel, left: schematic archtiecture of a bactosensor in which a glucose responsive promoter is coupled with an amplifying digital switch. In the presence of glucose, the recombinase is expressed and activates the switch. Upper panel, right: Insulation of bactosensors in stable hydrogel beads. Left: Uninduced beads. Right:Glucose-induced beads with RFP reporter. Lower panel: Operability of amplifying digital switches encapsulated in PVA/alginate beads. Beads that contained cells cotransformed with the XOR-RFP gate and pCpxP-TP901 were incubated in culture medium supplemented with various concentrations of glucose, and RFP fluorescence was measured after 24 hours. RFU, relative fluorescence unit. Detection thresholds for urinary dipsticks and for bactosensor are indicated. Pictures of the beads at various inducer concentrations are shown. Scale bar, 0.5 cm. (C) Bactosensormediated detection of abnormal glycosuria levels in clinical samples from diabetic patients. PVA/alginate beads encapsulating cells transformed with both the pCpxP-TP901 controller and the XOR-GFP gate, XOR-GFP gate alone, or the reference construct J23100-GFP were incubated in urine samples. Left panel: three glucose-negative samples from independent individuals and three positive controls [same samples supplemented with1%(w/v) glucose]. Right panel: Urine samples from 10 nonstabilized individual diabetic patients. GFP fluorescence was measured after 24 hours. Response to glucose was compared with standard urinary dipsticks (lower panels). The lower panels show the glucose reactive band of the urinary dipstick. In the absence of glucose, the band is blue. When glucose is present, the band turns brown. Data from three different experiments performed on different days are depicted by black, gray, and white circles. Each circle corresponds to one replicate (three replicates for each experiment).
Amplifying Genetic Logic Gates
Jérôme Bonnet, Peter Yin, Monica E. Ortiz, Pakpoom Subsoontorn and Drew Endy. "Amplifying genetic logic gates". Science, 2013, May 3;340(6132):599-603.
One major goal of current synthetic biology research is to "program" living cells to respond specifically to certain combinations of intra-or extra-cellular signals. In electronics, complex computation is achieved by combining elementary logic gates and applying Boolean algebra. For example, an AND gate gives a signal output only when its two input signals are on. Here we developed an original approach to engineering logic gates allowing us to "program" living cells like tiny computers.
Boolean integrase logic gates use two unidirectional integrases to control the expression of a gene of interest according to a specific combination of transcriptional inputs. As in an electronic transistor which control the flow of current through the circuits, the gates we developed control the flow of RNA polymerase along DNA. Logical functions can be simply programmed via the DNA sequence by changing the arrangement of recombination sites. we engineered a family of logic gates recapitulating all elementary boolean logic functions (AND, NAND, OR, NOR, XOR, XNOR) in single E. coli cells.
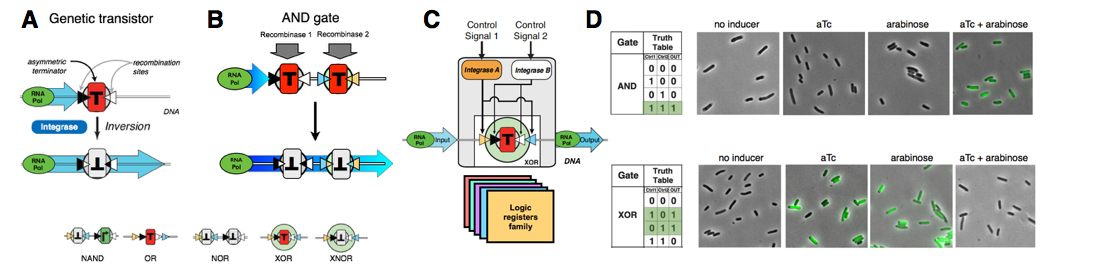
Mechanism and operation of Boolean integrase logic gates. (A) Transcriptor-based gates use integrases control signals to modulate RNA polymerase flow between a separate gate input and output. A canonical transcriptor element wherein an asymmetric transcription terminator (T) blocks transcriptional current in one orientation (red) or, when flipped, the opposite orientation (gray). Opposing recombination sites (black/white triangles) flank the terminator and direct flipping by an integrase. Bottom: Various genetic architectures encoding the different Boolean Logic functions.(B) A recombinase AND gate. Two integrases control inversion of a DNA target sequence flanked by the respective recombination sites. Each target contains a transcription terminator blocking transcription coming from a 5' promoter. To obtain an output from the AND gate, the two terminators must be inverted by the two integrases. (C) schematic of a full logic gate: two integrases controlled by different signal control the switching of a logic register. The logic function can be simply changed by using a different register.(D) Examples of AND and XOR gates operation in E. coli. Integrases are activated in response to anhydrotetracycline (aTc) or arabinose (ara), and gates activation produce the Green Fluorescent protein GFP
Rewritable digital data storage in live cells via engineered control of recombination directionality.
Jerome Bonnet, Pakpoom Subsoontorn and Drew Endy. PNAS, 2012, 109(23):8884-8889
The use of synthetic biological systems in research, healthcare, and manufacturing often requires autonomous history-dependent behavior and therefore some form of engineered biological memory. For example, the study or reprogramming of aging, cancer, or development would benefit from genetically encoded counters capable of recording up to several hundred cell division or differentiation events. Here we engineered a rewritable biological data storage systems reliably storing digital information within the chromosomes of living cells. Using recombinases, it is possible to invert a DNA sequence flanked by two recombination sites.
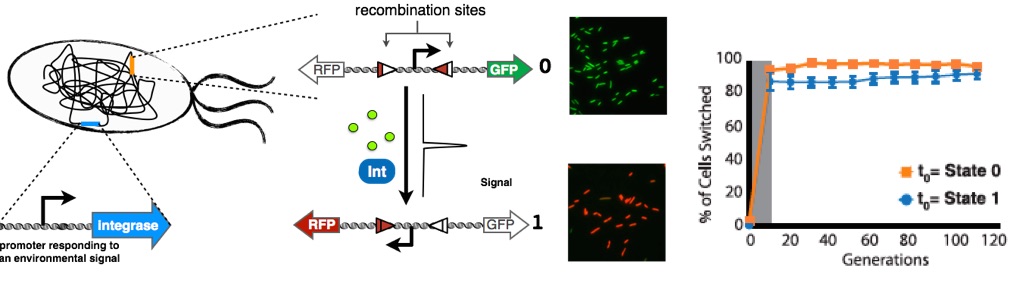
Operation of an integrase switch. The integrase is placed under the control of an inducible promoter (responding to arabinose). A recombination target, in which a constitutive promoter is flanked by recombination sites is placed on another part of the chromosome. In state 0 the promoter drives GFP expression. When integrase expression is induced, the promoter is flipped and drives RFP. Importantly, the reaction is unidirectional, resulting in data storage of the inducing signal over many generations even after inducer removal (right panel, gray area represents period of induction).
Thus, depending on the orientation of this sequence, two states can be stored in a DNA molecule, comparable to an electronic memory unit or "bit" (for binary digit). In electronics, rewritable bits allow the construction of binary counters which capacity increases exponentially (2ⁿ where n is the number of bits), enabling a large number of states with less components. However, no tools that allowing to reliably and reversibly write information within DNA in vivo had been engineered so far.
The RAD (Recombinase Addressable Data storage) module that we built in E. coli use the integrase and excisionase of the Bxb1 bacteriophage to reversibly and repeatedly invert a DNA sequence located between two recombination sites. Among the recombinase family of DNA-modifying enzymes, phage integrases are unique in that recombination directionality can be controled by an excisionase cofactor. We optimized the relative synthesis and degradation rates of both proteins by combining standard parts, directed evolution, screening and selection methods. The RAD module can store its state and remains functional for more than 100 cell divisions without performance degradation. These re-writable biological "bits" can be used to create new tools to study and control biological systems for basic research and biotechnology (counters and other systems requiring sequential logic).
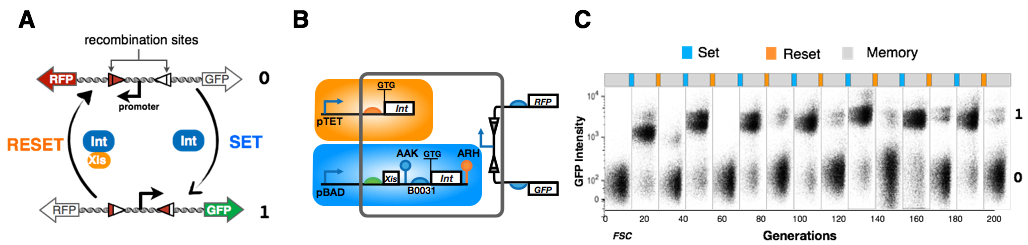
Rewritable digital data storage in live cells A) Principle of the RAD module based on DNA inversion by the integrase / excisionase system. Inversion of a constitutive promoter drives 2 different reporter genes, GFP and RFP. coexpression of integrate an excisions can reset the system tomatoes original state. (B) Details of the RAD module. Specific genetic regulatory elements controlling protein synthesis and degradation were obtained from standard biological parts collections, via computational design, or via random mutagenesis and screening. pTet promoter, inducible by anhydrotetracycline), controls transcription of integrase, while the promoter pBAD, induced by arabinose, drives simultaneous transcription of the integrase and excisionase (C) Experimental RAD module operation over multiple cycles in response to pulses of aTc (SET, Blue) and arabinose (RESET, Orange) across 140 cell divisions. For each point; pulse 4H, Storage (no inducers,gray )16H.

