Fragment-Based Drug Design
J.-F. Guichou, M. Gelin, G. Labesse, R. Rahimova
The group is developing fragment-based drug design using X-ray crystallography, biophysics, chemo-informatics and soft/bio-compatible chemistry for therapeutics applications in virology, oncology and infectious diseases.
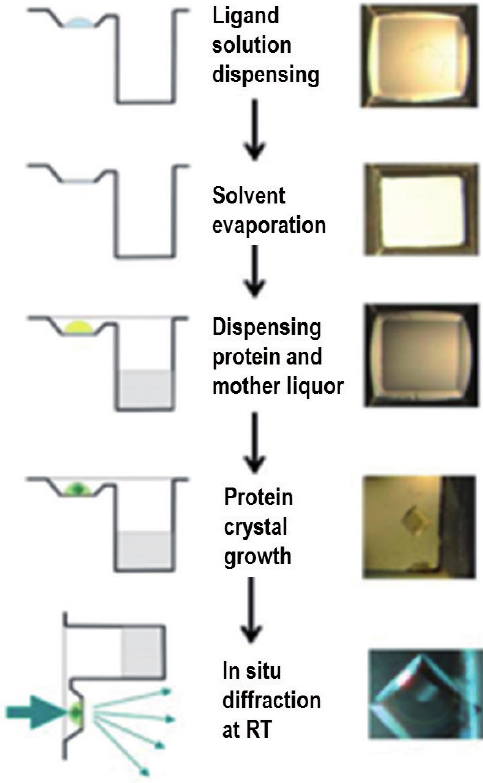
Designing new drugs remains highly challenging and complex with potential failure at each step. The group is developing an integrated approach for quicker and more rational design of drugs. Based on our expertise in ligand screening by X-ray crystallography, we have established a fully robotized platform for crystallization, crystal growth survey and diffraction in 96-well plates. This set-up is being used for the screening of fragment or more elaborate compounds generate by soft and/or bio-compatible chemistry. The group had also developed a fully automatic treatment for the resolution of the complex small molecule-protein (~150 structures per months). Combining all the structural information and the biophysics characterization (TSA, ITC, ...), the group design new therapeutic compounds which are tested through collaborations for their biological effects.
Collaborations : S. Pochet (I. Pasteur). I. Krimm (CNRS), JM. Pawlotsky (Hôpital Henri-Mondor). S. Roche (CNRS), P. Santa Barbara (INSERM), M. McGee (Dublin).
References : Gelin et al., Structure, 2012. Gelin et al., Acta Cryst D, 2015. Sagnol et al., NAR, 2015. Ahmed-Belkacem et al., Nat. Comm., 2016.
Fine enzymology and target deep-characterization
C. Lionne, M. Gelin, G. Labesse
The goal of the group is to help the design of efficient and specific drugs by studying enzyme reaction pathways and drug-target interactions by the mean of fine enzymology, biophysical characterization and thermodynamic approaches.
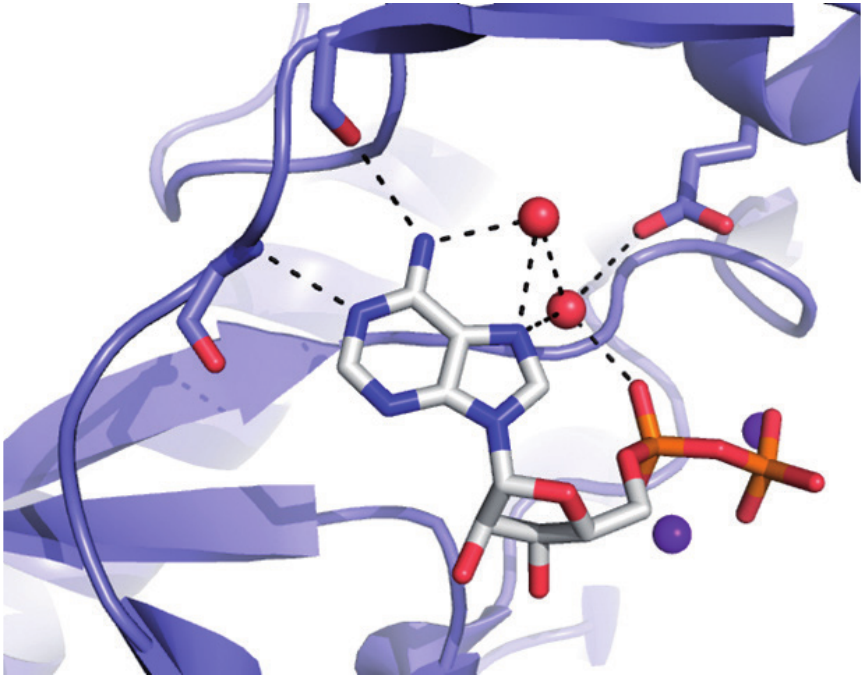
Bacterial antibiotic modifying enzyme: identification of the main reaction intermediate to be targeted.
Efficient and specific drugs directed against enzymes are much easier to design with a deep understanding of how the proteins work, and especially in which states they spend most of their time. Transient kinetics techniques (stopped-flow, quench-flow, cryoenzymology) are routinely used to measure protein-ligand kinetics (kon, koff ), identify reaction pathway and rate-limiting steps, as well as to characterize the effects of inhibitors thereon. Beside, other biophysical techniques are used to characterize drug-target interactions: classical steady state enzymology, isothermal titration calorimetry, thermal shift assay, molecular docking and X-ray crystallography. Optimized inhibitors are designed and their biological effects tested (in collaboration for cellular and animal models). The group focuses its research on new targets working on nucleos/tides for the design of innovative anti-infectious or anti-cancer therapies.
Collaborations: Zohra Befodda (CHROME Nîmes), Anne Blanc-Potard, Rachel Cerdan & Sharon Wein-Gratraud (LPHI Montpellier), Konstantin Brodolin & Laurent Chaloin (IRIM, Montpellier), Vincent Chaptal (IBCP Lyon), Catherine Dunyach-Remy (VBIC Nîmes), Felix Hausch (Technische Universität Darmstadt), Gerta Hoxhaj (University of Texas Dallas), Jean-Michel Jault et Elise Kaplan (IBCP Lyon), Thierry Lorca (CRBM Montpellier), Suzanne Peyrottes & Christophe Mathé (IBMM Montpellier), Sylvie Pochet & Olivier Dussurget (Institut Pasteur Paris), Katharina Rox (HZI Braunschweig).
Recent references: Praharaj et al. Sci Adv. in press; Kaplan et al. ChemMedChem. 2025; Lacroix et al. EMBO J. 2024; Morichaud et al. Nat Commun. 2023; Clément et al. Eur J Med Chem. 2023; Rahimova et al. FEBS J. 2023; Mary et al. Mol Cell. 2022.