-
Postdoctoral position – Team Integrative Biophysics of Membranes
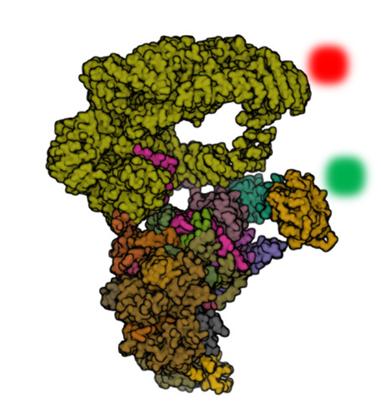
A 2 year postdoctoral position is available in single molecule fluorescence spectroscopy and protein engineering, to study eukaryotic transcription initiation.
Supervisor : E. Margeat.
- 1
Stages

Le CBS accueille chaque année de nombreux stagiaires de tout niveau, de la 3ème au post-doctorat. Les candidatures (CV et lettre de motivation) sont à adresser directement aux chercheurs des équipes du CBS.
Le recrutement des étudiants en Thèse se fait par l'intermédiaire de l'Ecole Doctorale CBS2.
Les sujets de Thèse proposés par les chercheurs du CBS sont à consulter ici.
PROGRAMME DE STAGES DE SECONDE – Cliquer ici

