Permanent staff of the team involved in platform projects: P. Barthe, K. de Guillen, A. Padilla, C. Roumestand
Team members are in charge of the PIBBS NMR platform which carries out numerous punctual or long-term studies in collaboration with groups from the CBS and other institutes in Montpellier, in France or abroad. Many of the 3D structures solved by the team are the result of such collaborative projects, notably the M. Tuberculosis Structural Biology project.
Platform projects leading to publications:
2019
. NMR structure of an effector protein of the rust fungus Melampsora larici-populina: MLP124017
. Solution structure and backbone dynamics of LSR2 and its phosphomimetic LSR2-T112D from Mtb
. Backbone chemical shift assignments of the corepressor N-CoR
2018
. NMR study of the interaction between FhaA and the PknB-phosphorylated cytoplasmic CwlM from Mtb
. NMR analysis of the tumor-targeting molecule FAPI-02
. Towards the total synthesis of trichormamide A, a cyclic undecapeptide
2016
. NMR Structure of the Mycobacterium tuberculosis LppM
. Volume analysis of Hsp90 protein-ligand binding site by high-pressure NMR
2015
. TGAM_1934, an ORFan protein from the archeon Thermococcus gammatolerans
. NMR analysis of the external PASTA domain of PknB from Mycobacterium tuberculosis
2014
. NMR structure of an Antimicrobial peptide
2013
. NMR structure of the adhesion protein EDK-∆-Bd37 from Babesia divergens
. Fragment based drug discovery – NMR screening by STD
. NMR structure of an highly specific tumor-targeting miniprotein
(2012 and before) Other past projects
2019
NMR structure of an effector protein of the rust fungus Melampsora larici-populina: MLP124017
NMR Platform Staff : K. de Guillen, P. Barthe & A. Padilla
Protein Size : 149 a.a. 15N and 13C labeled
Rust fungi are plant pathogens that secrete an arsenal of effector proteins interfering with plant functions and promoting parasitic infection. We successfully purified and solved the three-dimensional structure, using NMR spectroscopy, of MLP124017 an effector of the leaf rust fungus Melampsora larici-populina in Escherichia coli. It exhibits structural similarities to nuclear transport factor 2-like proteins, which are molecular containers involved in a wide range of functions
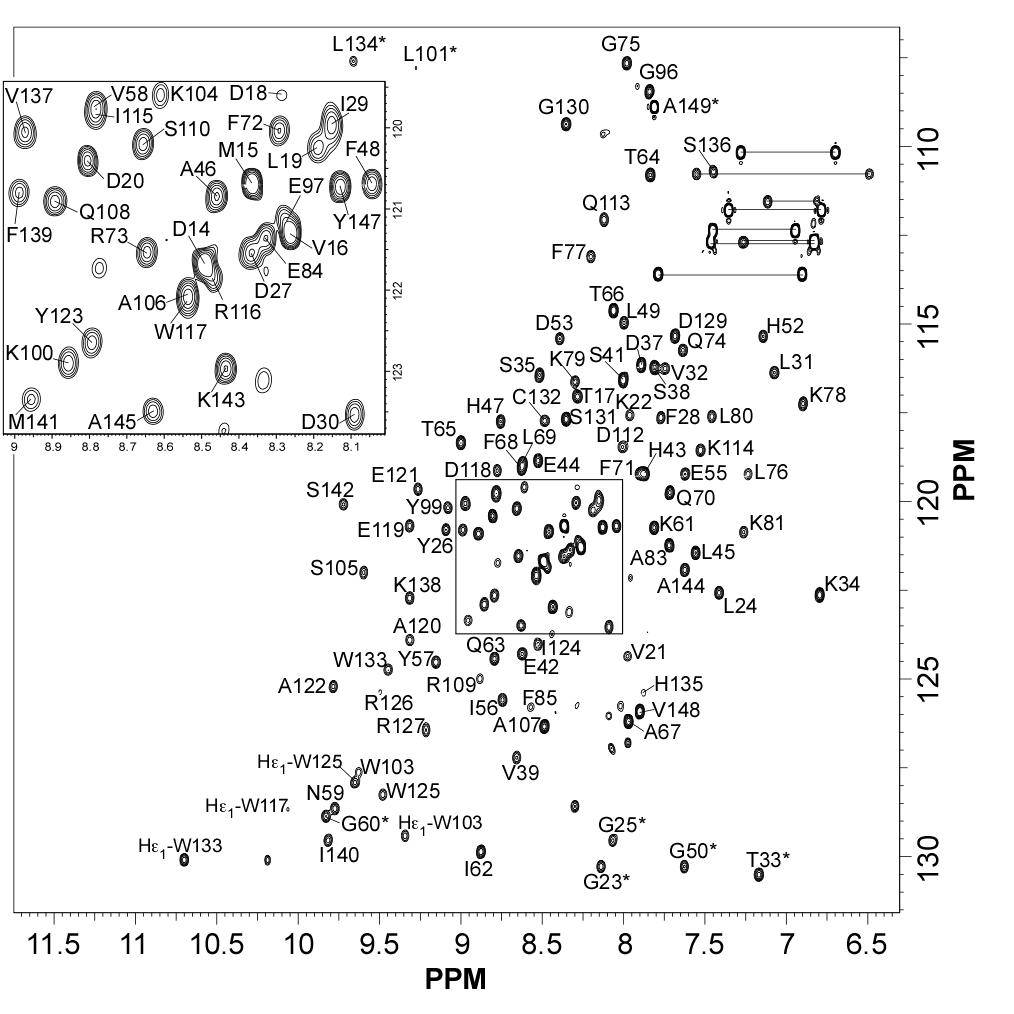 |
 |
| [1H-15N]-HSQC of MLP124017 | Ribbon 3D structure of the MLP124017 protein. beta-sheet are represented by purple arrows, helices are coloured in green. |
External Contact: A. Hecker (Université de Lorraine, INRA, IAM, Nancy)
De Guillen K, Lorrain C, Tsan P, Barthe P, Petre B, Saveleva N, Rouhier N, Duplessis S, Padilla A & Hecker A. Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds. Sci Rep. 2019 Dec 2;9(1):18084. doi: 10.1038/s41598-019-53816-9.
Solution structure and backbone dynamics of LSR2 and its phosphomimetic LSR2-T112D from Mtb
NMR Platform Staff : P. Barthe & C. Roumestand
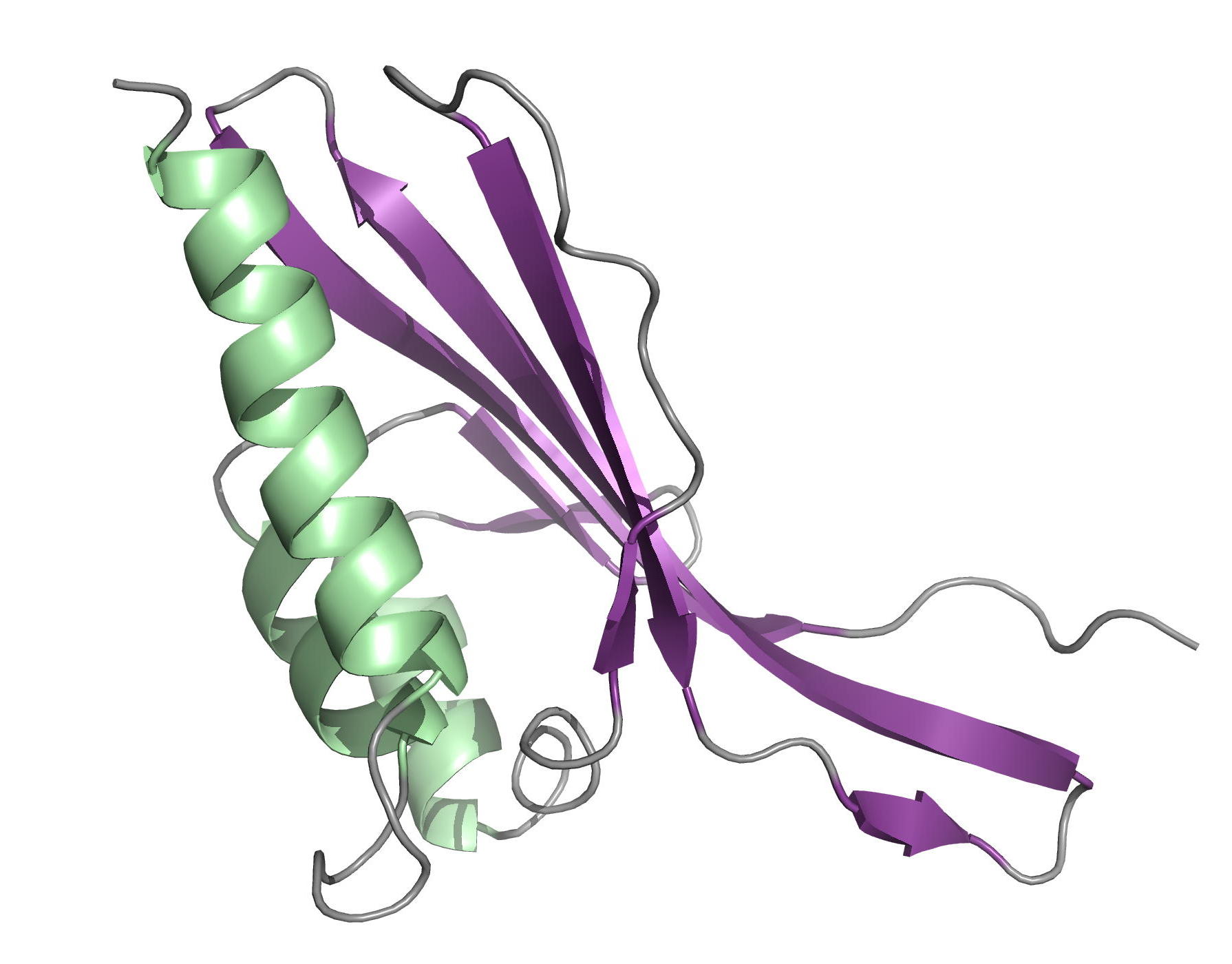
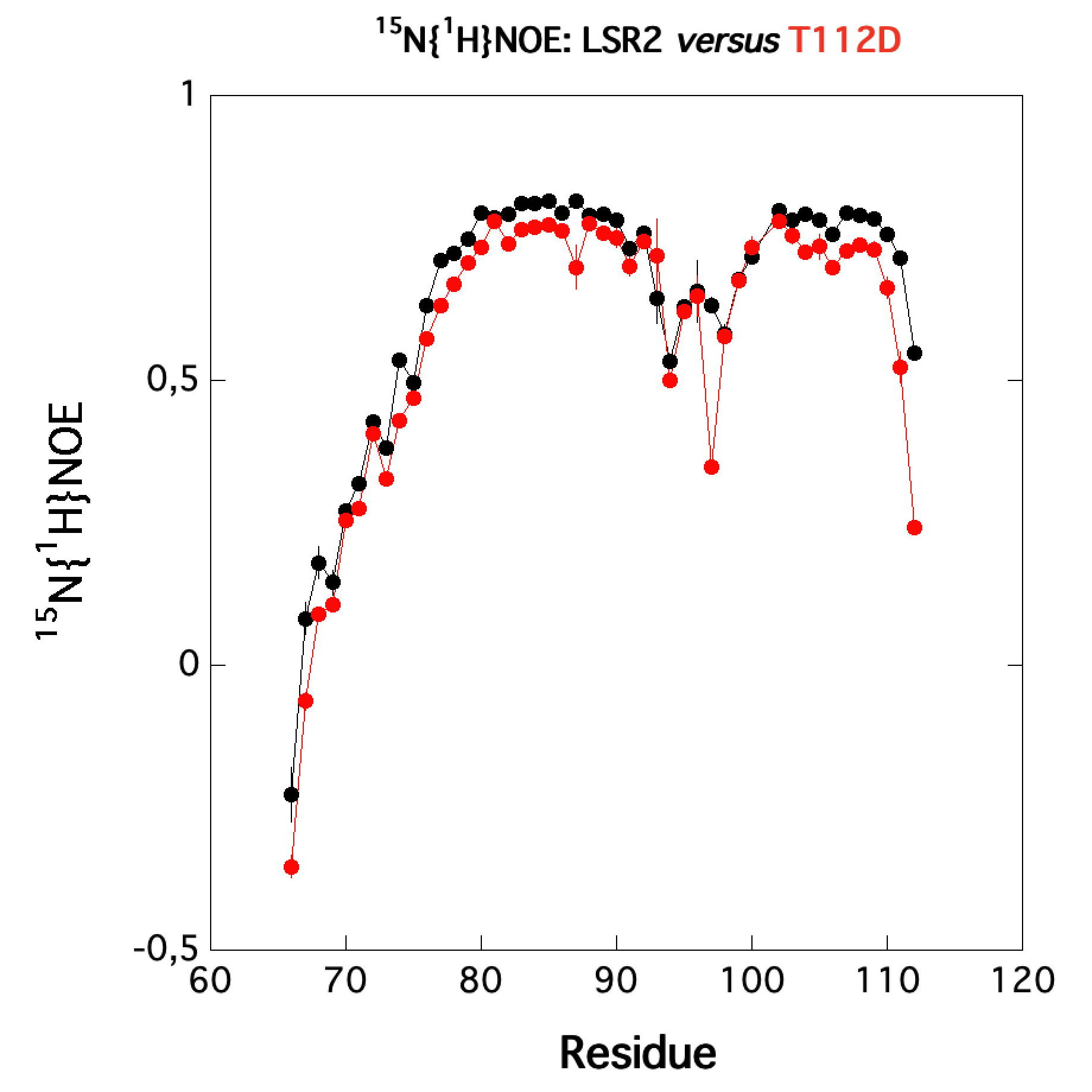
Protein kinase B (PknB) controls peptidoglycan biosynthesis of Mycobacterium tuberculosis (Mtb) during growth. The kinase phosphorylates the H-NS-like regulator Lsr2 at threonine 112. PknB phosphorylation of Lsr2 reduces DNA binding, measured by fluorescence anisotropy and electrophoretic mobility shift assays, and our NMR structure of phosphomimetic T112D Lsr2 suggests that this may be due to increased dynamics of the DNA-binding domain.
 |
 |
| Comparison of the heteronuclear nOe profiles of LSR2 (in black) and LSR2-T112D (in red) | Ribbon 3D structure of the LSR2 peptide (in green) and its phosphomimetic LSR2-T112D (in red) |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Alqaseer K, Turapov O, Barthe P, Jagatia H, de Vish A, Roumestand C, Wegrzyn M, Bartek I, Voskuil M, O'Hare H, Ajuh P, Bottrill A, Witney A, Cohen-Gonsaud M, Waddell S & Mukamolova G. Protein kinase B controls Mycobacterium tuberculosis growth via phosphorylation of the transcriptional regulator Lsr2 at threonine 112. Mol Microbiol. 2019 Dec;112(6):1847-1862. doi: 10.1111/mmi.14398.
Backbone chemical shift assignments of the corepressor N-CoR
NMR Platform Staff : P. Barthe.
Protein Size : 269 a.a. 15N and 13C labeled
In complex with the retinoic X receptor (RXR), the retinoic acid receptor (RAR) regulates the transcription of numerous genes. This activity is facilitated by the recruitment of transcriptional co-repressors in the promoter region of the target genes. Unexpectedly we found that, while mainly disordered, the corepressor N-CoR presents evolutionary conserved structured regions involved in transient intramolecular contacts.
| [1H-15N]-HSQC of N-CoR protein (269 aa) | 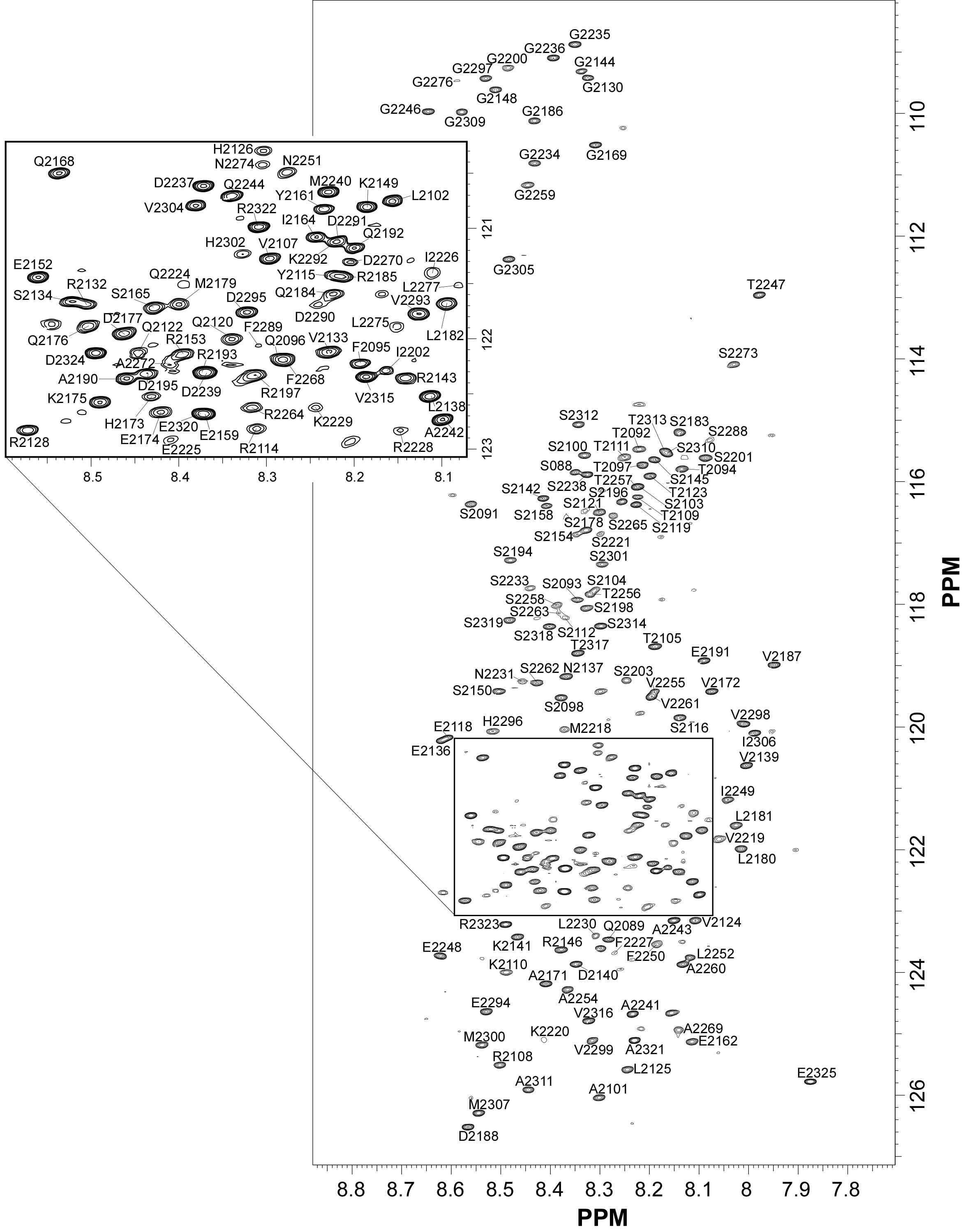 |
Internal Contact: P. Bernado or N. Sibille (Centre de Biochimie Structurale, Montpellier)
Cordeiro T, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, Bourguet W & Bernado P. Interplay of protein disorder in retinoic acid receptor heterodimer and its corepressor regulates gene expression. Structure. 2019 Aug 6;27(8):1270-1285.e6. doi: 10.1016/j.str.2019.05.001.
2018
NMR study of the interaction between FhaA and the PknB-phosphorylated cytoplasmic CwlM from Mtb
NMR Platform Staff : P. Barthe
Protein kinase B (PknB) phosphorylates CwlM, an essential regulator of peptidoglycan synthesis in Mycobacterium tuberculosis (Mtb). We demonstrate that growing mycobacteria produce two forms of CwlM: a non-phosphorylated membrane-associated form and a PknB-phosphorylated cytoplasmic form. We show that the partner proteins for the phosphorylated and non-phosphorylated forms of CwlM are FhaA, a fork head-associated domain protein, and MurJ, a proposed lipid II flippase, respectively.
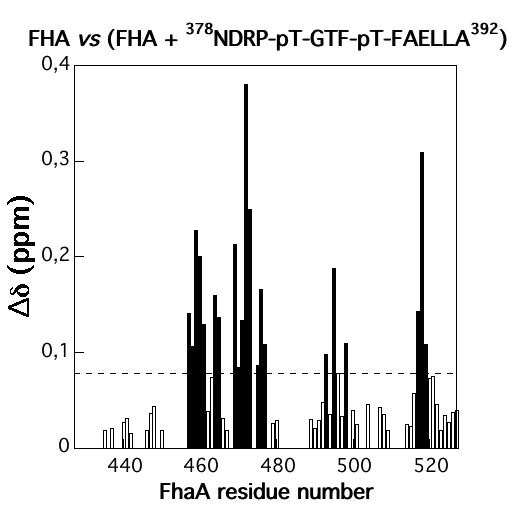 |
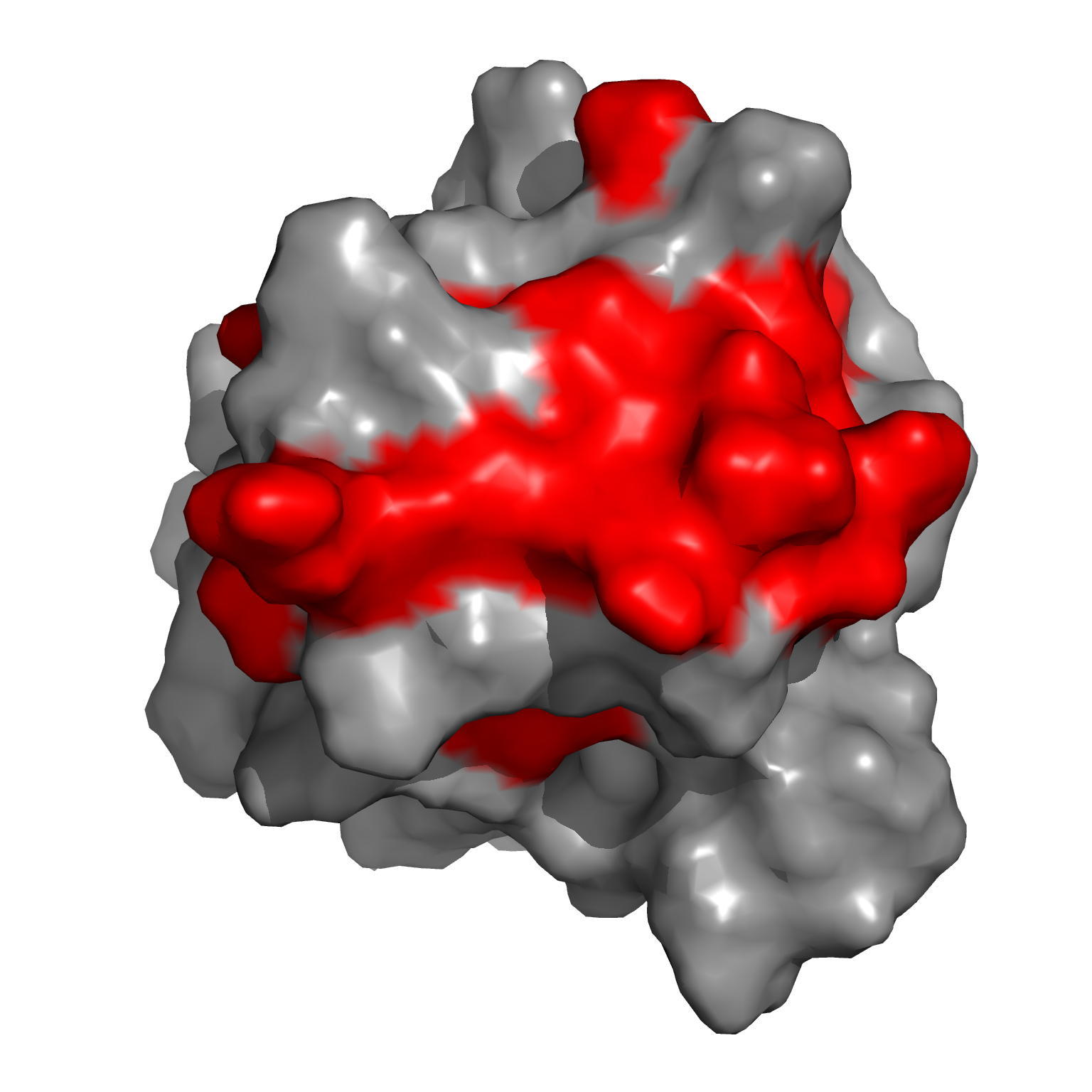 |
| Amide averaged chemical shift variations (∆∂) as a function of protein sequence | Chemical shift perturbations by CwlM-peptides on the surface of Rv0020c-FHA |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Turapov O, Forti F, Kadhim B, Ghisotti D, Sassine J, Straatman-Iwanowska A, Bottrill A, Moynihan P, Wallis R, Barthe P, Cohen-Gonsaud M, Ajuh P, Vollmer W & Mukamolova G. Two faces of CwlM, an essential PknB substrate, in Mycobacterium tuberculosis. Cell Rep. 2018 Oct 2;25(1):57-67.e5. doi: 10.1016/j.celrep.2018.09.004.
NMR analysis of the tumor-targeting molecule FAPI-02
NMR Platform Staff : P. Barthe & C. Roumestand
The tumor stroma, which accounts for a large part of the tumor mass, represents an attractive target for the delivery of diagnostic and therapeutic compounds. Cancer-associated fibroblasts feature high expression of fibroblast activation protein (FAP), which is not detectable in adult normal tissue but is associated with a poor prognosis in cancer patients. Our collaborators developed a DOTA-coupled radiotracer based on a FAP-specific enzyme inhibitor (FAPI) and we verified it in vitro by NMR.
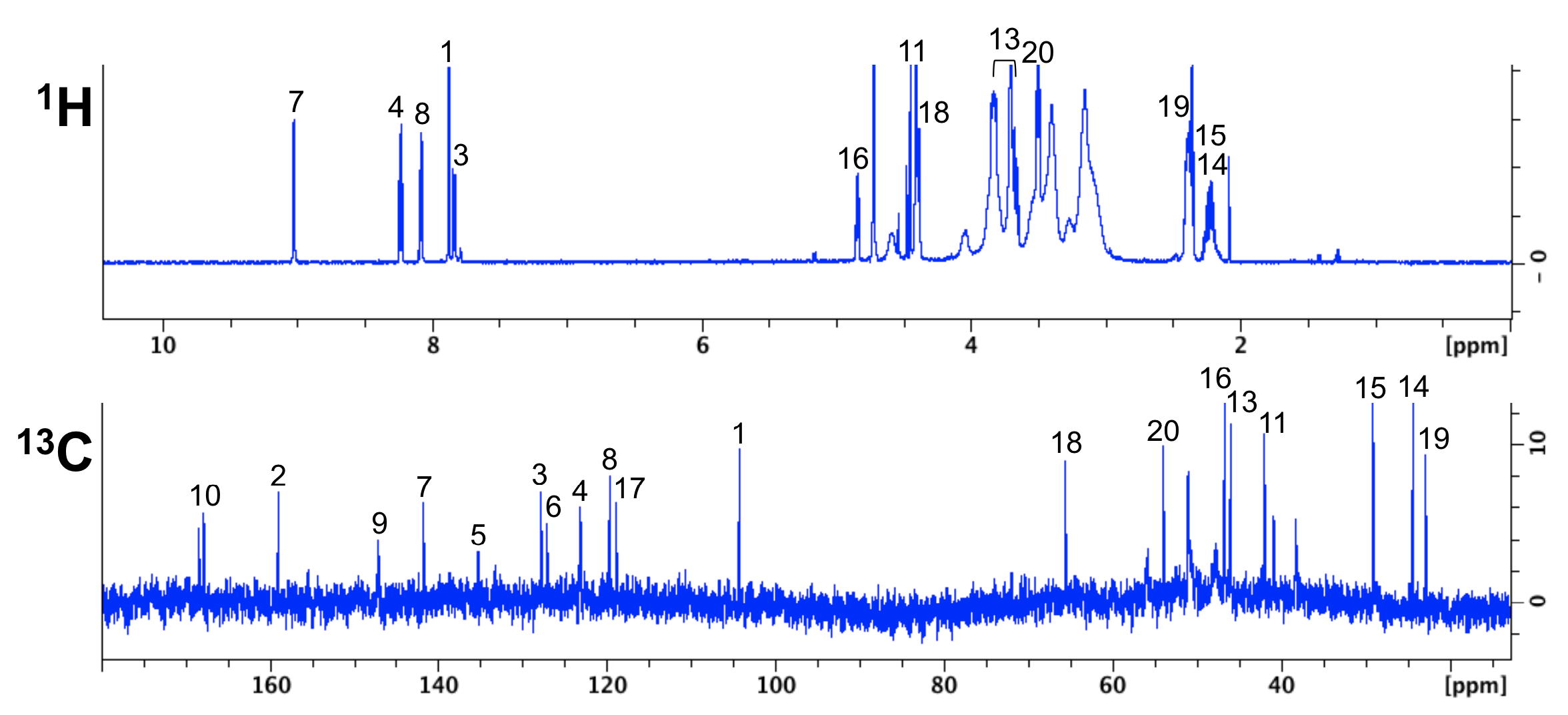 |
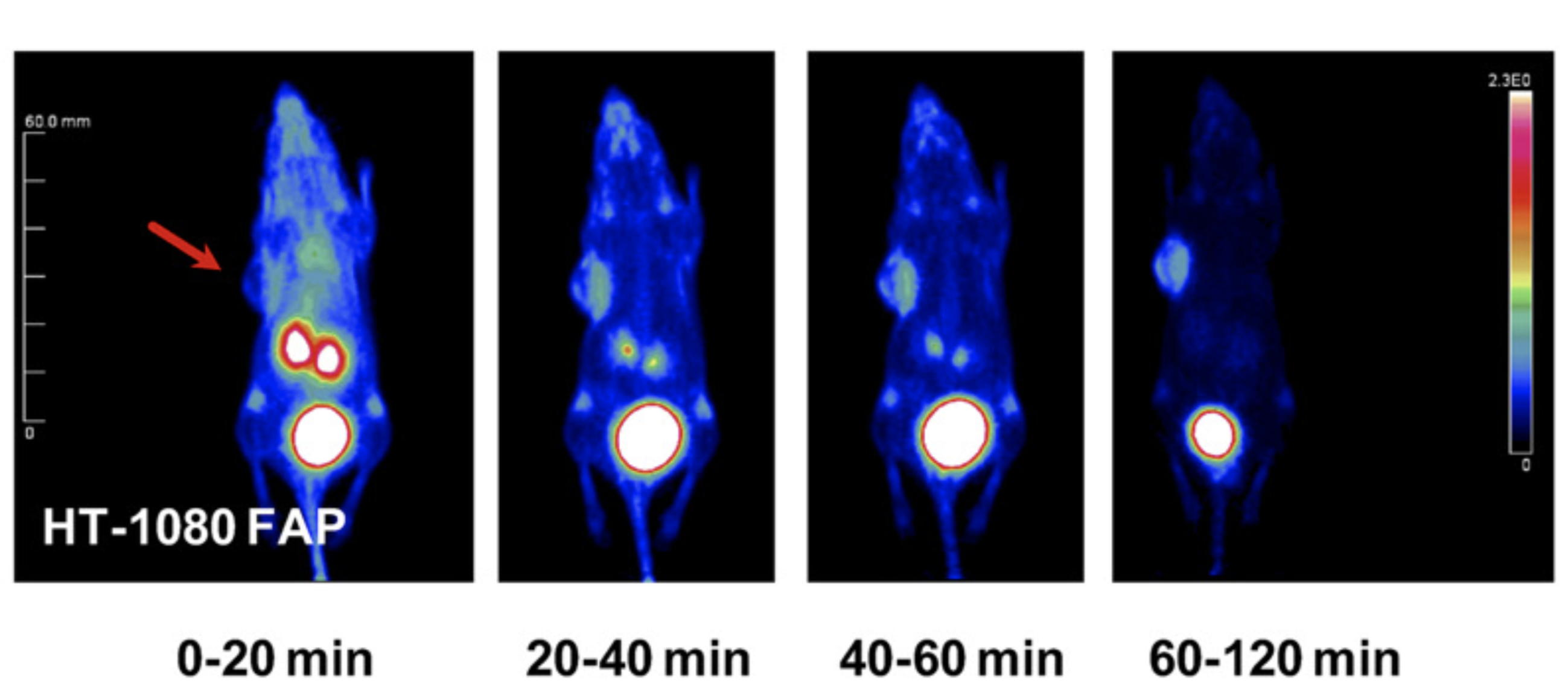 |
| NMR chemical shifts (1H and 13C) of the FAPI-02 molecule | 68Ga-FAPI-02 PET in mice bearing human FAP-expressing (HT-1080-FAP) xenografts |
External Contact: U. Haberkorn (University of Heidelberg, Germany)
Loktev A, Lindner T, Mier W, Debus J, Altmann A, Jager D, Giesel F, Kratochwil C, Barthe P, Roumestand C & Haberkorn U. A tumor-imaging method trageting cancer-associated fibroblasts. J Nucl Med. 2018 Sep;59(9):1423-1429. doi: 10.2967/jnumed.118.210435.
Towards the total synthesis of trichormamide A, a cyclic undecapeptide
NMR Platform Staff : C. Roumestand
Protein Lypocyclopeptides isolated from marine sources hold great promise as new lead for therapeutics. Here, we propose the first attempt towards the total synthesis of Trichormamide A. This synthesis help us to confirm the proposed structure of the cyclic lipopeptide.
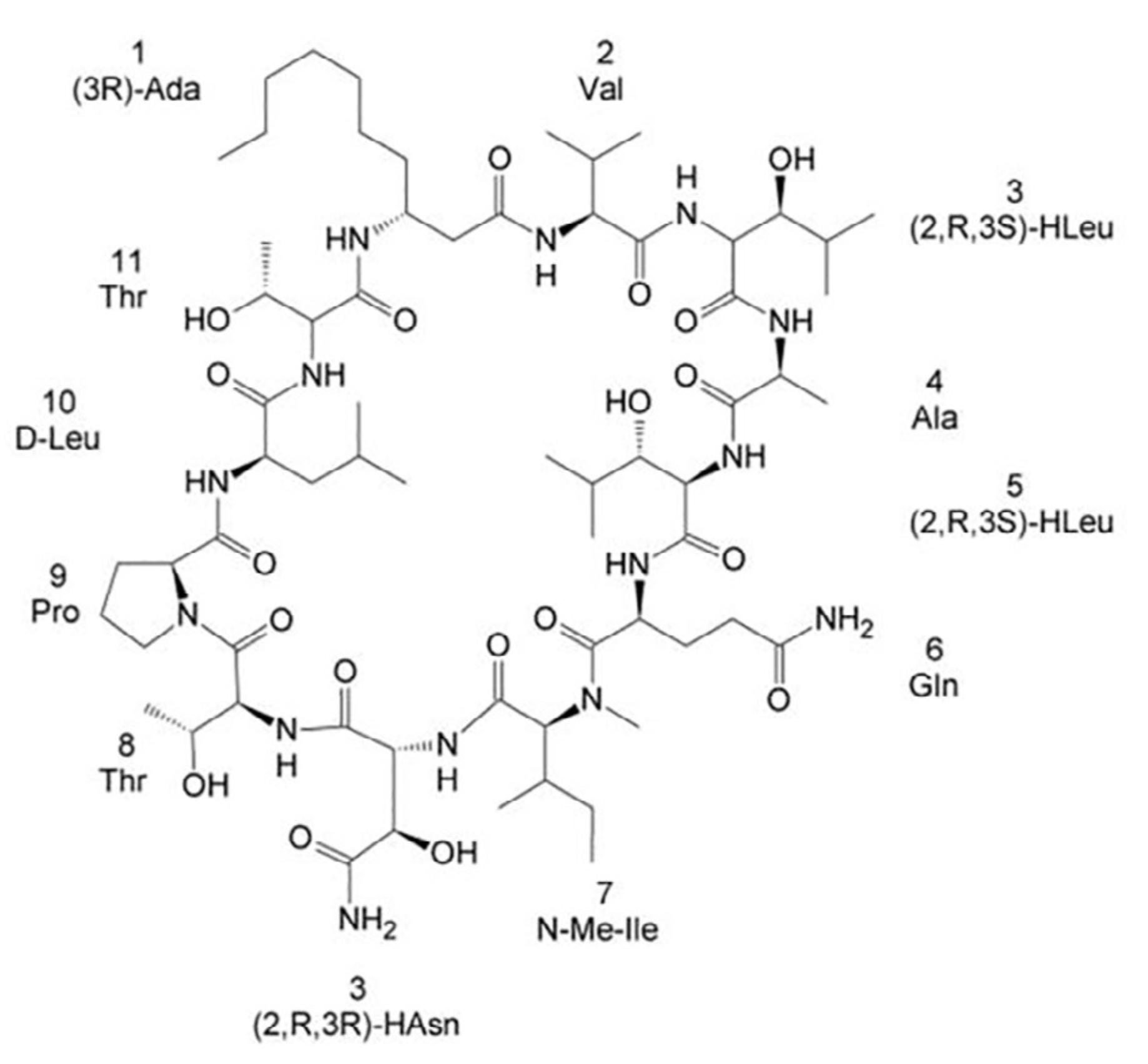 |
|
| Structure of Laxaphycin B |
External Contact: N. Inguimbert (USR 3278 CRIOBE, Perpignan)
Gaillard M, Das S, Djibo M, Raviglione D, Roumestand C, Legrand B & Inguimbert N. Towards the total synthesis of trichormamide A, a cyclic undecapeptide. Tetrahedron Lett. 2018 Oct 10;59(41):3713-3718. doi: 10.1016/j.tetlet.2018.09.010.
2016
NMR Structure of the Mycobacterium tuberculosis LppM
NMR Platform Staff : P. Barthe
Protein Size : 171 a.a. 15N and 13C labeled
LppM (Rv2171), a Mycobacterium tuberculosis (Mtb) protein, is both implicated in phagocytosis and able to efficiently block phagosomal acidification in the macrophage, two key processes contributing to Mtb persistence. We show that LppM is anchored to the mycobacterial cell wall by a C-terminal membrane domain. However, the protein also exists as a truncated protein secreted into the culture medium. The LppM solution structure displays no similarity with other Mtb lipoproteins.
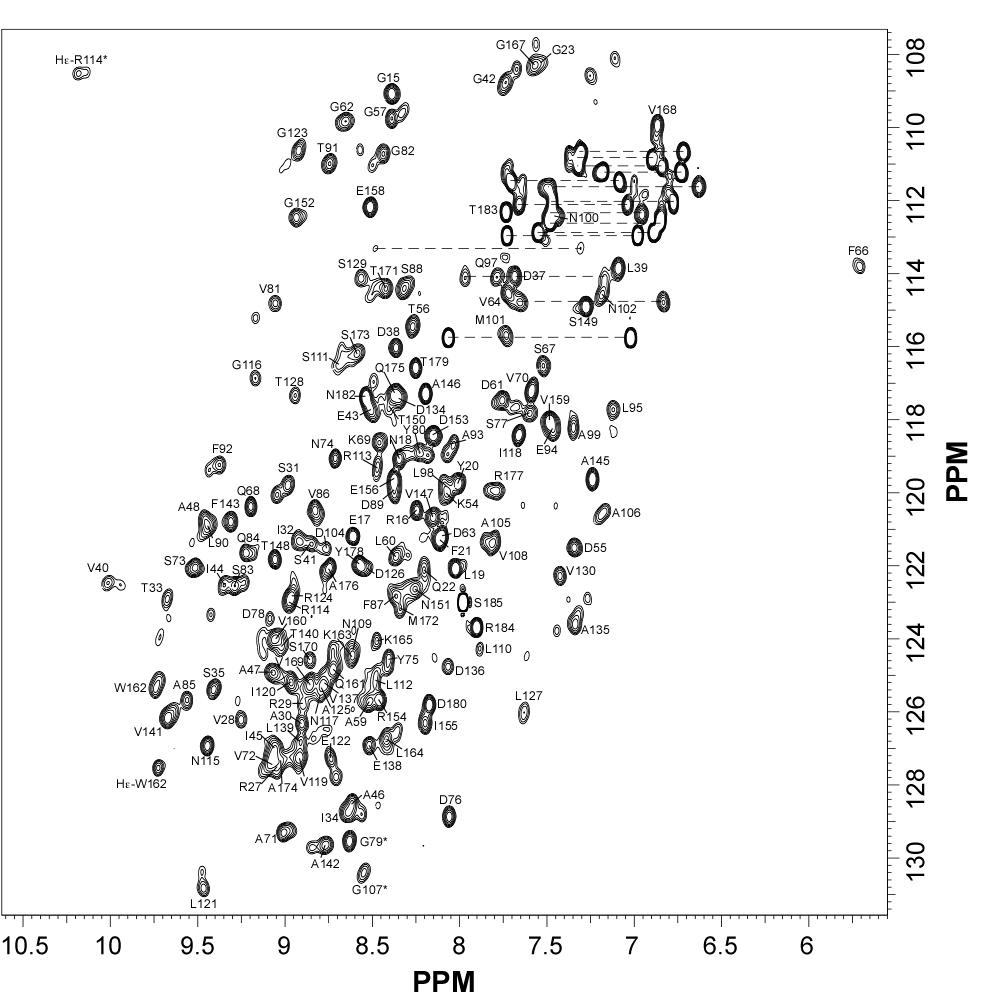 |
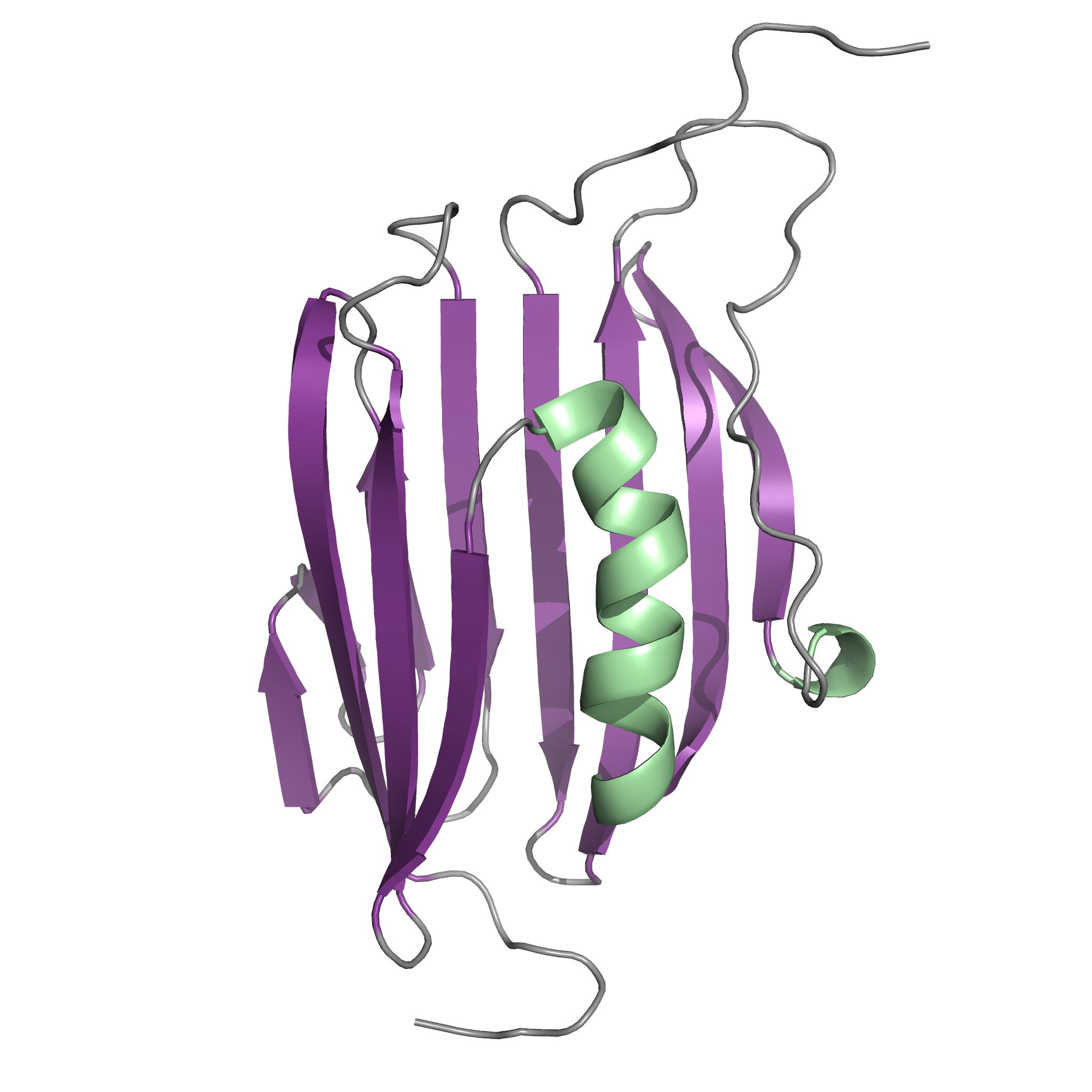 |
| [1H-15N]-HSQC of LppM (Rv2171) | Ribbon 3D structure of the LppM protein |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Barthe P, Veyron-Churlet R, de Visch A, Gilleron M, Saliou J-M, Tomavo S, Nigou J, Brodin P & Cohen-Gonsaud M. Mycobacterium tuberculosis LppM Displays an Original Structure and Domain Composition Linked to a Dual Localization. Structure. 2016 Oct 4;24(10):1788-1794. doi: 10.1016/j.str.2016.07.009.
Volume analysis of Hsp90 protein-ligand binding site by high-pressure NMR
NMR Platform Staff : C. Roumestand
Human Heat Shock Protein 90 (Hsp90) is a key player in the homeostasis of the proteome and plays a role in numerous diseases, such as cancer. The volume of inhibitor binding is an important parameter to understand the relationship between an inhibitor structure and its inhibition potential. Here, the volumes of binding of several ligands to recombinant Hsp90 were obtained by three independent experimental techniques: fluorescent pressure shift assay, vibrating tube densitometry, and high-pressure NMR. Within the error range, all techniques provided similar volumetric parameters for the investigated protein−ligand systems.
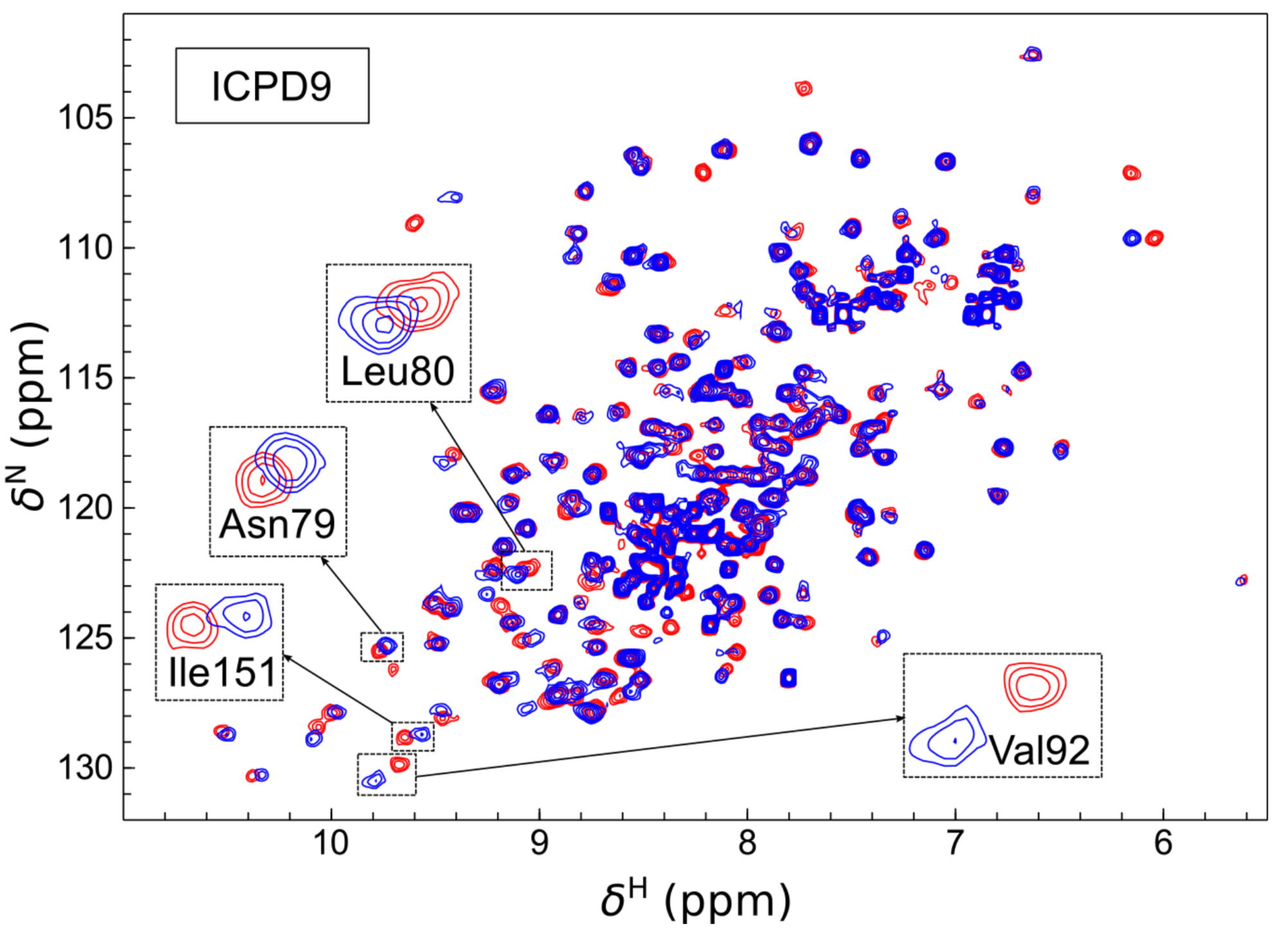 |
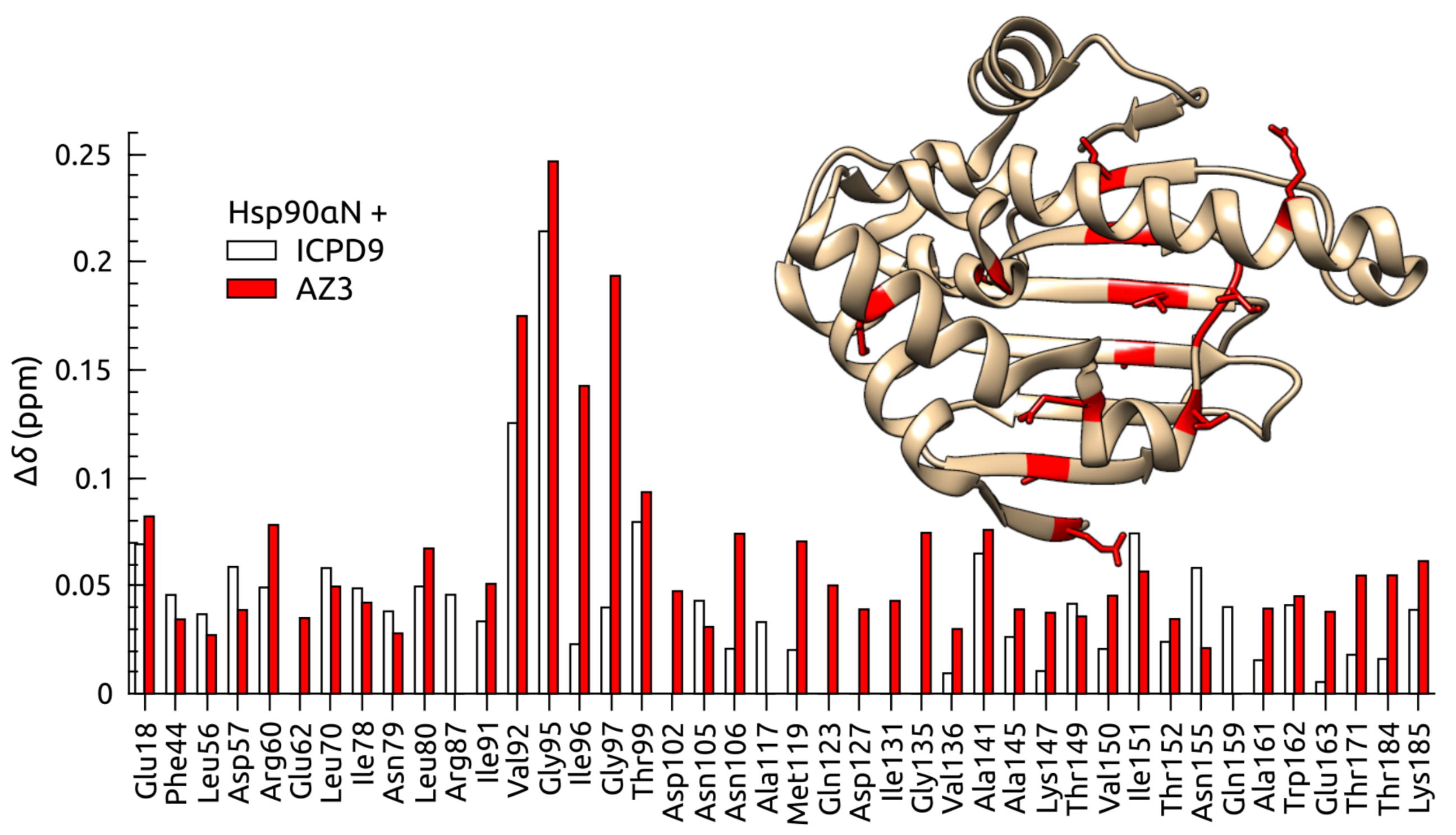 |
| [1H-15N]-HSQC of spectra of Hsp90N with (blue) and without (red) the ICPD9 ligand | Chemical shift changes of selected residues of Hsp90N after the addition of AZ3 (solid bars) and ICPD9 (open bars) ligands. |
External Contact: V. Petrauskas (University of Vilnius, Lithuania)
Toleikis Z, Sirotkin V, Skvarnavicius G, Smirnoviene J, Roumestand C, Matulis D & Petrauskas V. Volume of Hsp90 protein-ligand binding determined by fluorescent pressure shift assay, densitometry, and NMR. J Phys Chem B. 2016 Sep 22;120(37):9903-12. doi: 10.1021/acs.jpcb.6b06863.
2015
TGAM_1934, an ORFan protein from the archeon Thermococcus gammatolerans
NMR Platform Staff : K. de Guillen, Y. Yang & C. Roumestand
Protein Size : 148 a.a. 15N and 13C labeled
ORFans are proteins found when annotating new genomes by comparative genomics. TGAM_1934, a 122 amino acid polypeptide from the hyper-radioresistant T. gammatolerans archaeon, was selected as the most abundant hypothetical protein amongst the low molecular weight proteome. The NMR results highlight the potential of structural proteogenomics, i.e. prioritizing ORFan targets for structure determination based on quantitative proteomics data.
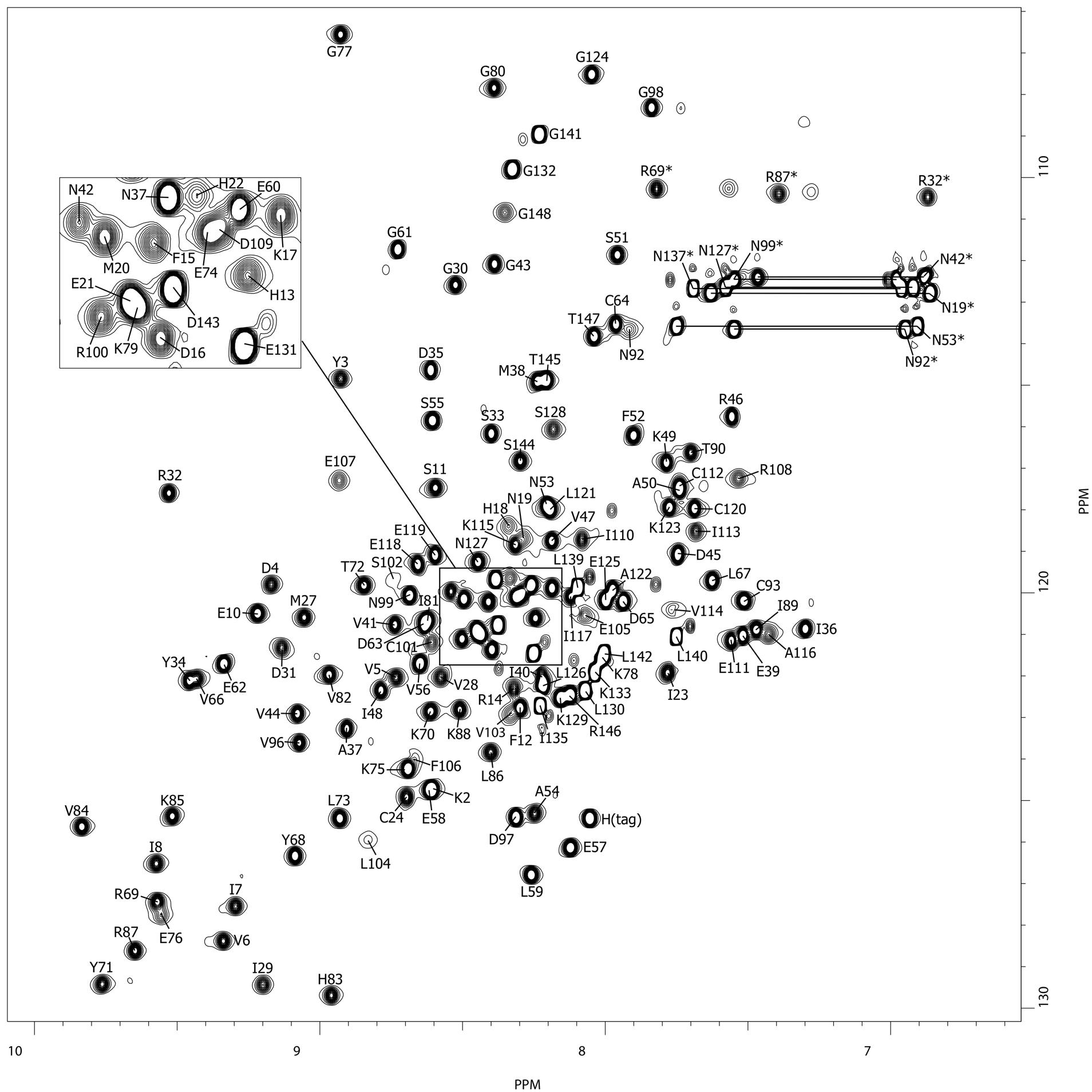 |
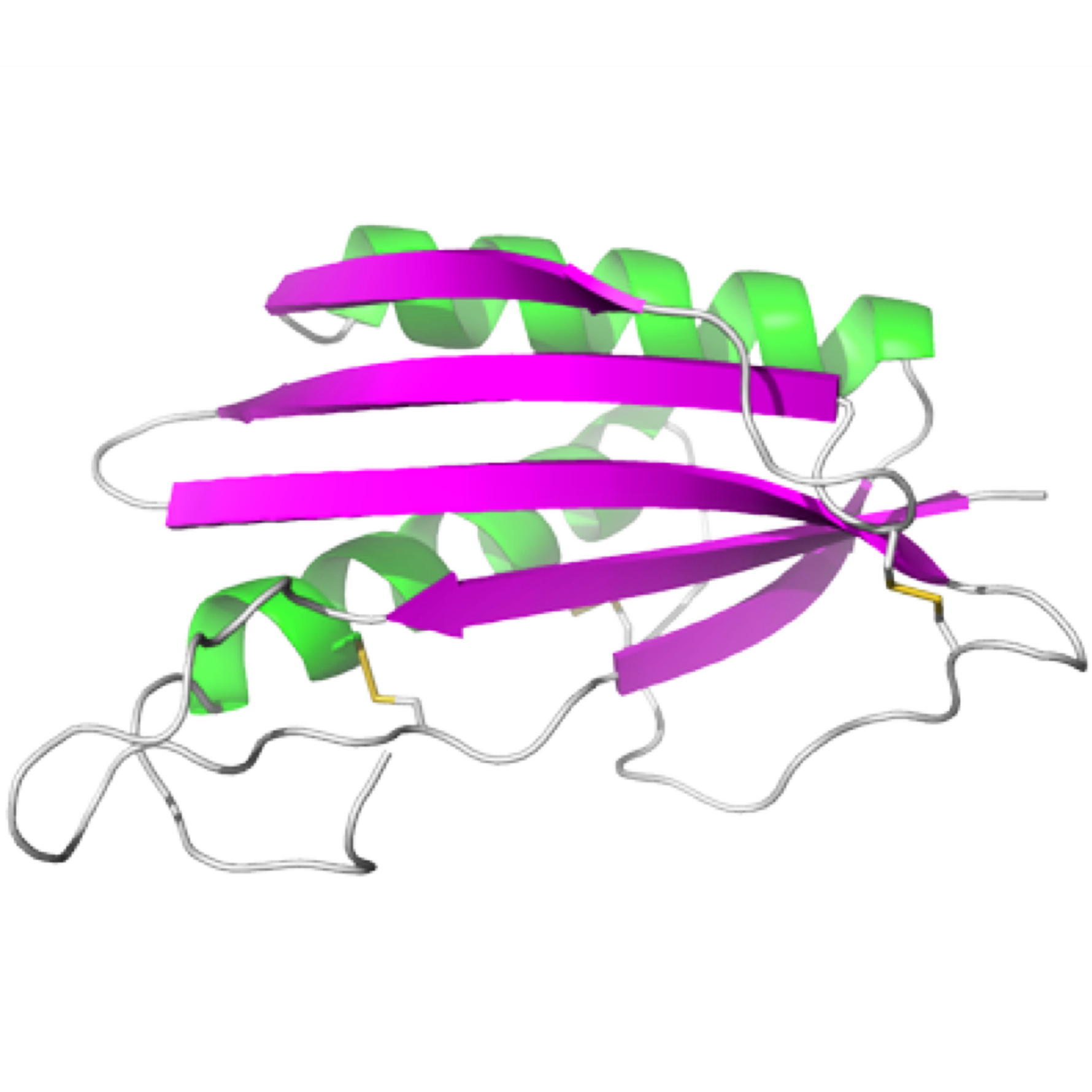 |
| [1H-15N]-HSQC of TGAM_1934 | Ribbon 3D structure of TGAM_1934. Beta-sheet is represented by purple arrows. |
External Contact : J. Armengaud (CEA, DSV, IBEB, Bagnols-sur-Cèze)
Yang Y, Fernandez B, Lagorce A, Aloin V, de Guillen K, Boyer J, Dedieu A, Confalonieri F, Armengaud J & Roumestand C. Prioritizing targets for structural biology through the lens of proteomics: the archaeal protein TGAM_1934 from Thermococcus gammatolerans, Proteomics. 2015 Jan;15(1):114-23. doi: 10.1002/pmic.201300535.
NMR analysis of the external PASTA domain of PknB from Mycobacterium tuberculosis
NMR Platform Staff : P. Barthe
PknB is an essential serine/threonine protein kinase required for mycobacterial cell division and cell-wall biosynthesis. We show that the addition of recombinant PASTA domain could prevent regrowth of Mycobacterium tuberculosis, and therefore offers an alternative opportunity to control replication of this pathogen. These results suggest that the PknB_PASTA domain is involved in regulation of peptidoglycan biosynthesis and maintenance of cell-wall architecture.
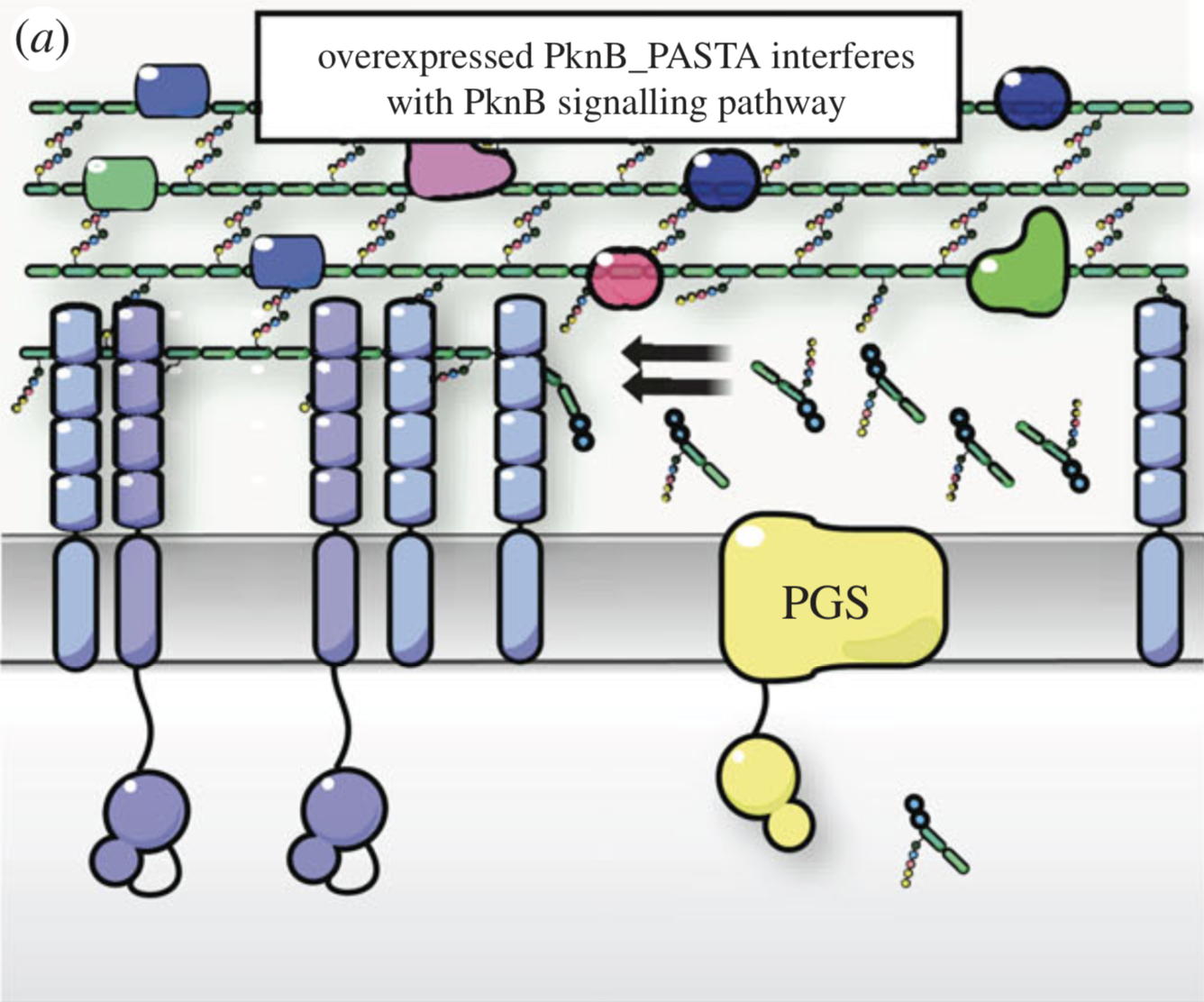 |
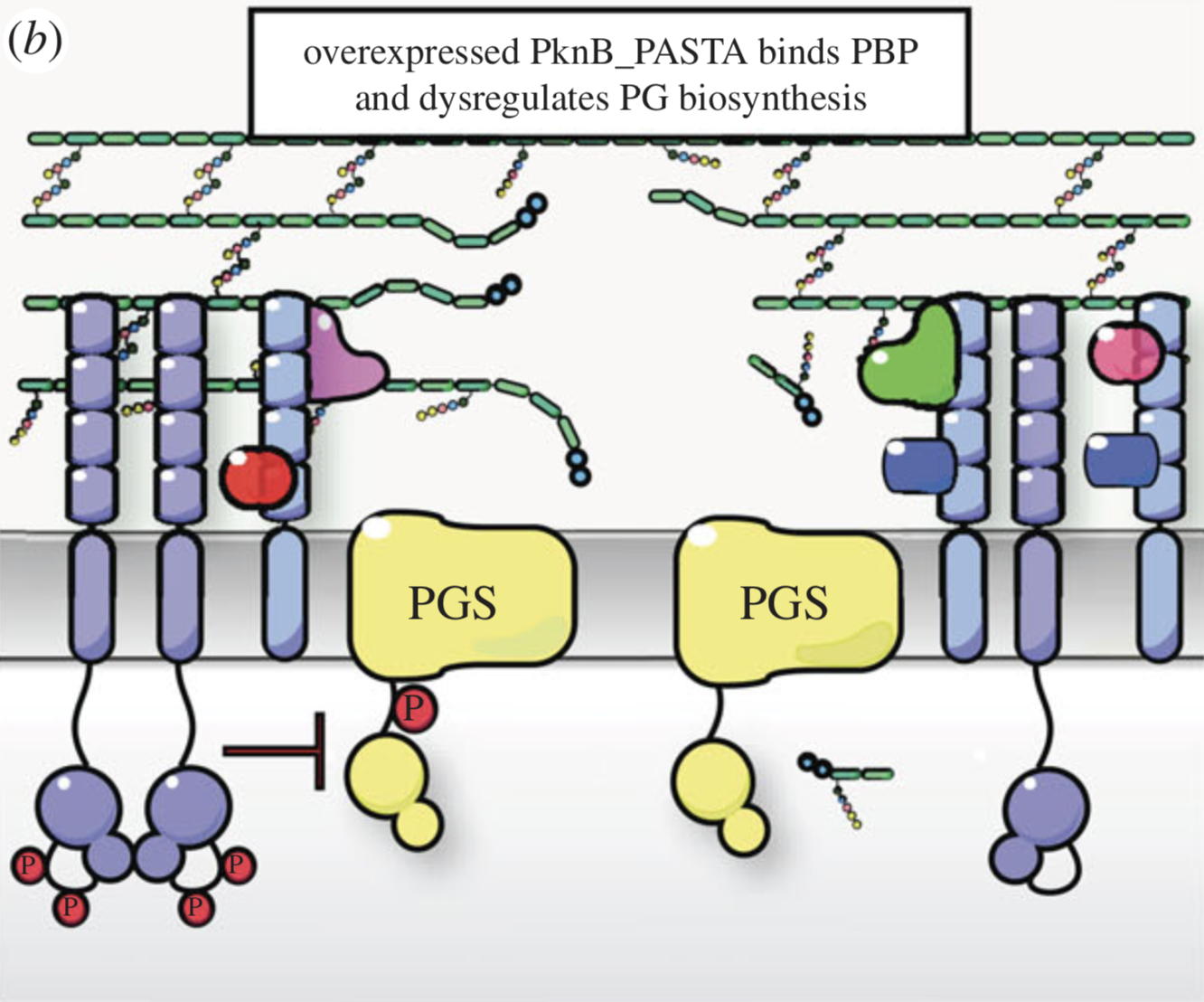 |
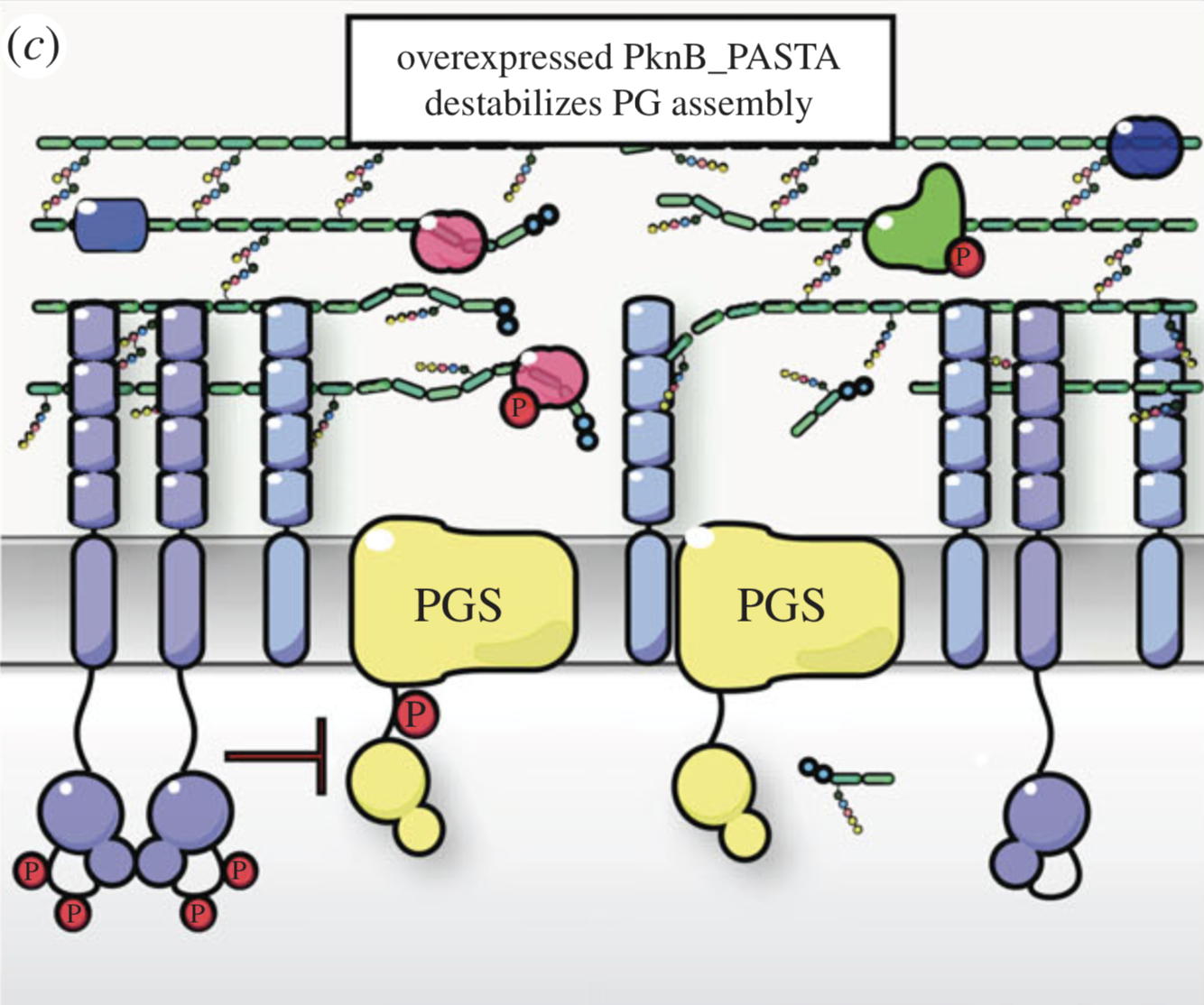 |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Turapov O, Loraine J, Jenkins C, Barthe P, McFeely D, Forti F, Ghisotti D, Hesek D, Lee M, Bottrill A, Vollmer W, Mobashery S, Cohen-Gonsaud M & Mukamolova G. The external PASTA domain of the essential serine/threonine protein kinase PknB regulates mycobacterial growth. Open Biol. 2015 Jul;5(7):150025. doi: 10.1098/rsob.150025.
2014
NMR structure of an Antimicrobial peptide
NMR Platform Staff : André Padilla (pipeline from CBS RX platform)
Peptide : GGLKKLGKKLEGAGKRVFKASEKALPVVVGIKAIGK Unlabeled
Aedesin is a cecropin-like anti-microbial peptide that was recently isolated from dengue virus-infected salivary glands of the Aedes aegypti mosquito. In the present study, we have refined the analysis of its structural characteristics. Aedesin has a helix-bend-helix structure typical for a member of the family of alpha-helix anti-microbial peptides.
 |
 |
| Spectra overlay : TOCSY (green) NOESY (yellow) | Surface coloured by the charge, blue for positive, red for negative and grey for hydrophobic residues. |
External Contact: D. Missé (MIVEGEC, UMR224 IRD/CNRS/UM, Montpellier)
Godreuil S, Leban N, Padilla A, Hamel R, Luplertlop N, Chauffour A, Vittecoq M, Hoh F, Thomas F, Sougakoff W, Lionne C, Yssel H & Misse D. Aedesin: Structure and antimicrobial activity against multidrug resistant bacterial strains. PLoS One. 2014 Aug 27;9(8):e105441. doi: 10.1371/journal.pone.0105441.
2013
NMR structure of the adhesion protein EDK-∆-Bd37 from Babesia divergens
NMR Platform Staff : P. Barthe & C. Roumestand
Protein Size : 224 a.a. 15N and 13C labeled
We report here the solution structure of EDK-∆-Bd37, a 25 kDa surface protein from the Apicomplexa parasite Babesia divergens that could undergo drastic conformational changes during erythrocyte invasion.
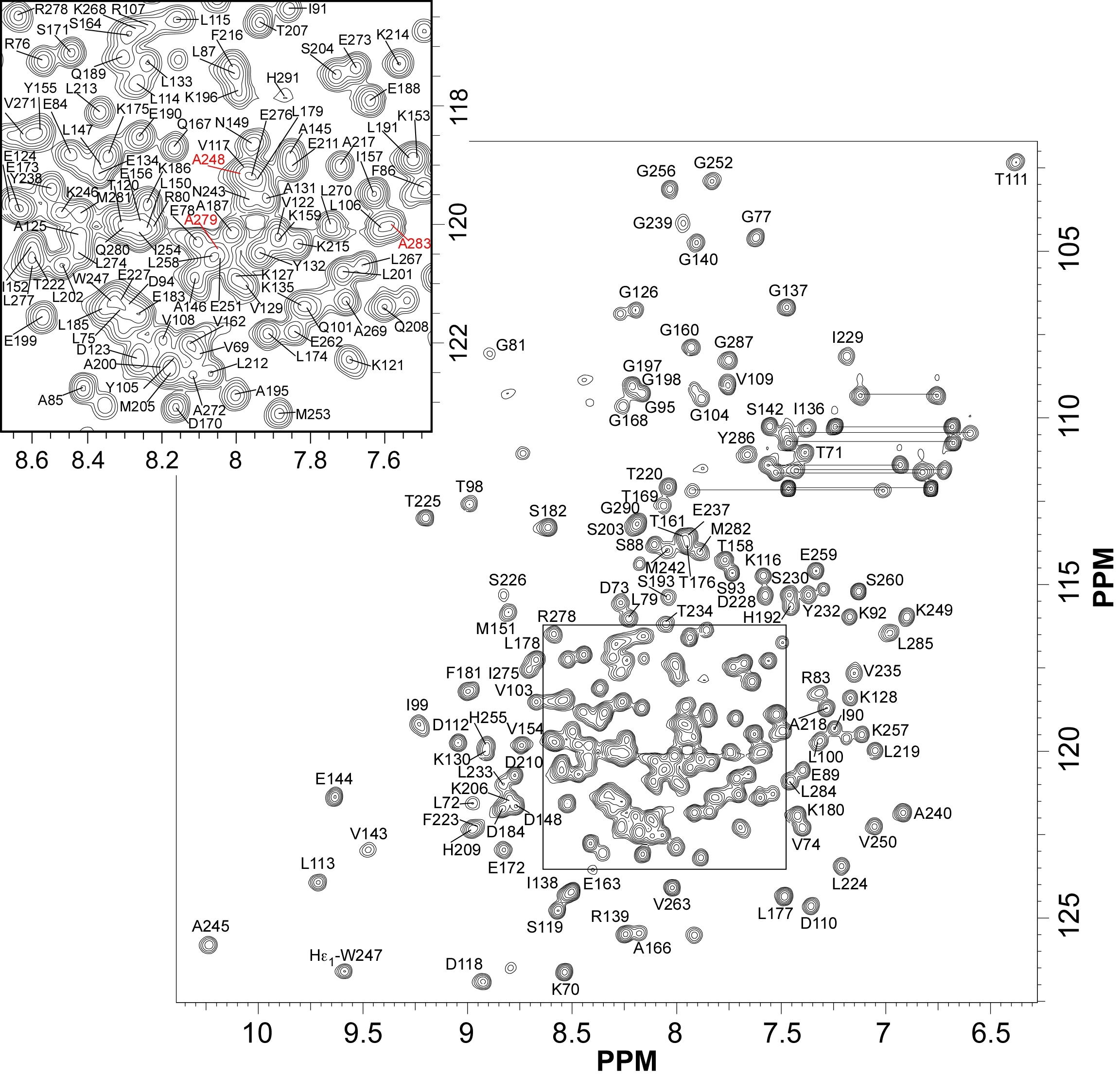 |
 |
| [1H-15N]-HSQC of EDK-∆-Bd37 | Ribbon 3D structure of the EDK-∆-Bd37 |
External Contact : Stéphane Delbecq (EA4558, Vaccination Antiparasitaire, Montpellier)
Barthe P, Murciano B, Schetters T, Gorenflot A, Delbecq S & Roumestand C. 1H, 15N and 13C backbone resonance assignments of a conformational mutant of the adhesion protein ∆-Bd37 from Babesia divergens. Biomol NMR Assign. 2013 Oct;7(2):241-4. doi: 10.1007/s12104-012-9418-6. Epub 2012 Aug 17.
Fragment based drug discovery – NMR screening by STD
NMR Platform Staff : André Padilla Pipeline to CBS RX platform
Protein Size: 22 kDa, Unlabeled @ 30 µM
By virtual screening using a fragment-based drug design (FBDD) approach, 33 fragments were selected within small pockets around interaction hot spots on the Sec7 surface of the nucleotide exchange factor Arno. By use of NMR, the direct binding of three of the identified fragments to Arno Sec7 domain was demonstrated and the promiscuous aggregate behavior evaluated.
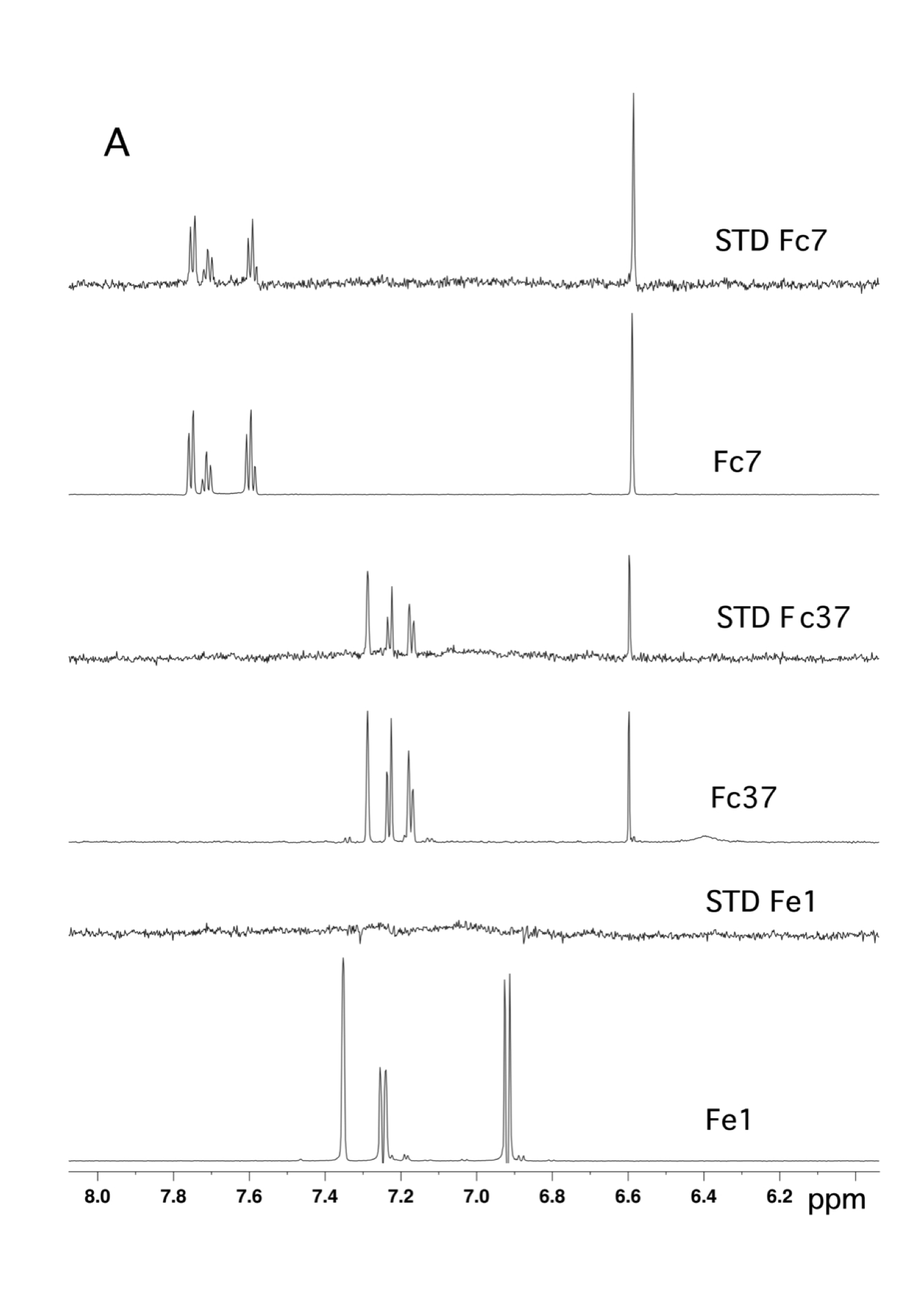 |
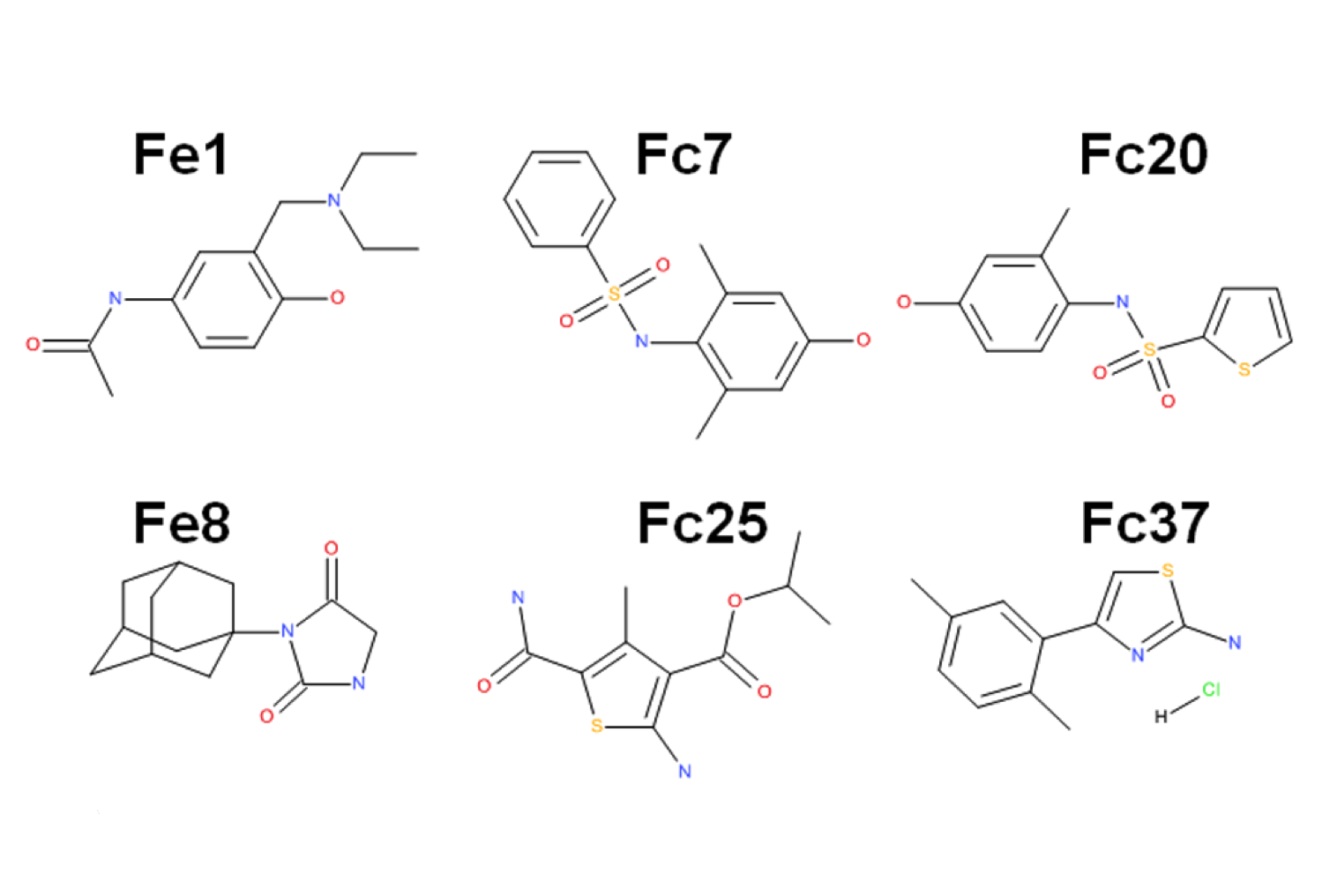 |
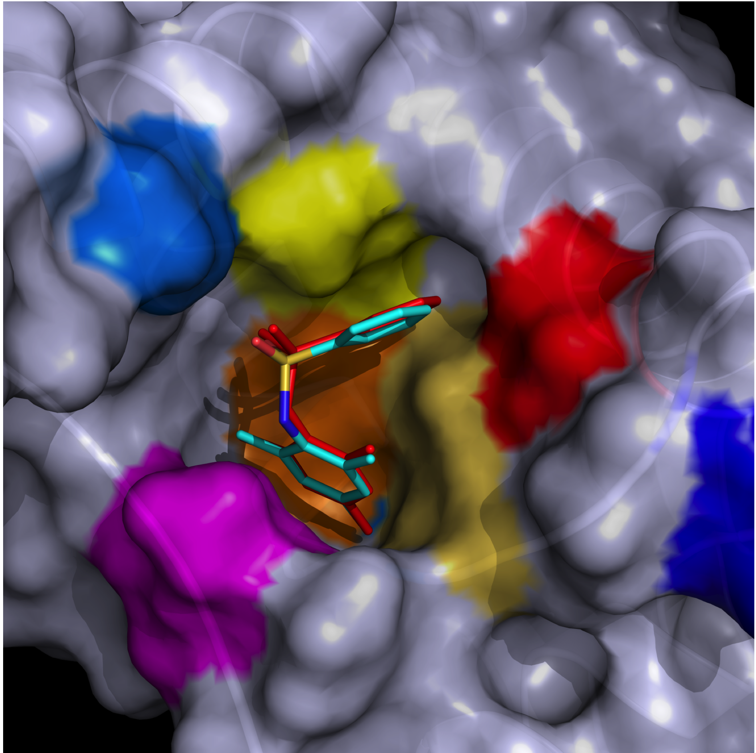 |
| STD spectra | XRay Structure of the ARNO-Sec7-FC7 complex |
External Contact: J. Rouhana (IBMM, Montpellier)
Rouhana J, Hoh F, Estaran S, Henriquet C, Boublik Y, Kerkour A, Trouillard R, Martinez J, Pugniere M, Padilla A & Chavanieu A. Fragment-based identification of a locus in the Sec7 domain of Arno for the design of protein-protein interaction inhibitors. J Med Chem. 2013 Nov 14;56(21):8497-511. doi: 10.1021/jm4009357. Epub 2013 Oct 29.
NMR structure of an highly specific tumor-targeting miniprotein
NMR Platform Staff : Philippe Barthe
In this study, the scaffold properties of the two-disulfide-stabilized miniprotein Min-23 (DLL-Rib) were exploited to generate new affinity reagents against the angiogenesis marker Delta-like Ligand 4 (DLL4) performing ribosome display. In both in vitro and in vivo studies it was shown, that only the properly folded miniprotein specifically bond to the target DLL4. These characteristics allowed for imaging of tumor with positron emission tomography applying radiolabeled miniproteins.
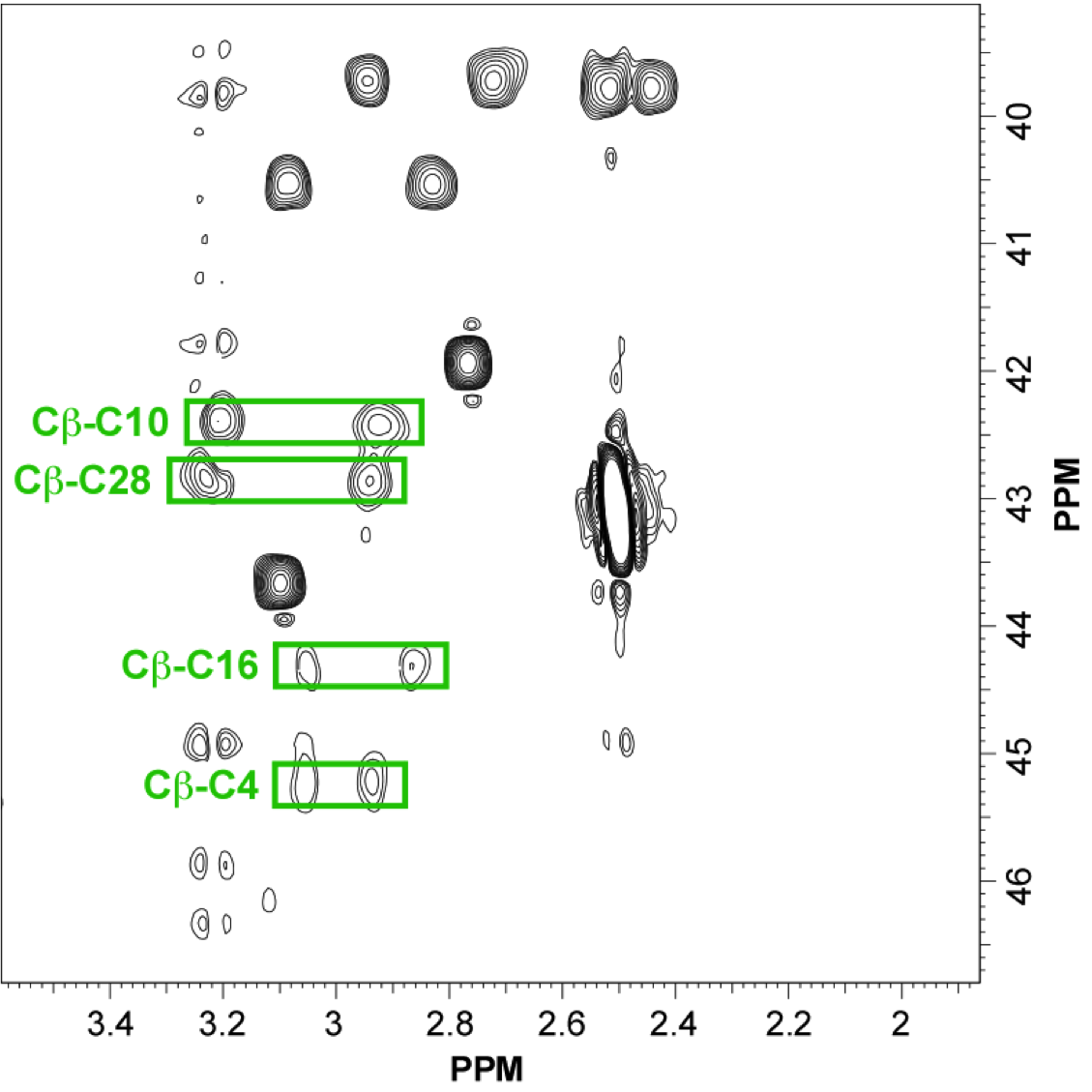 |
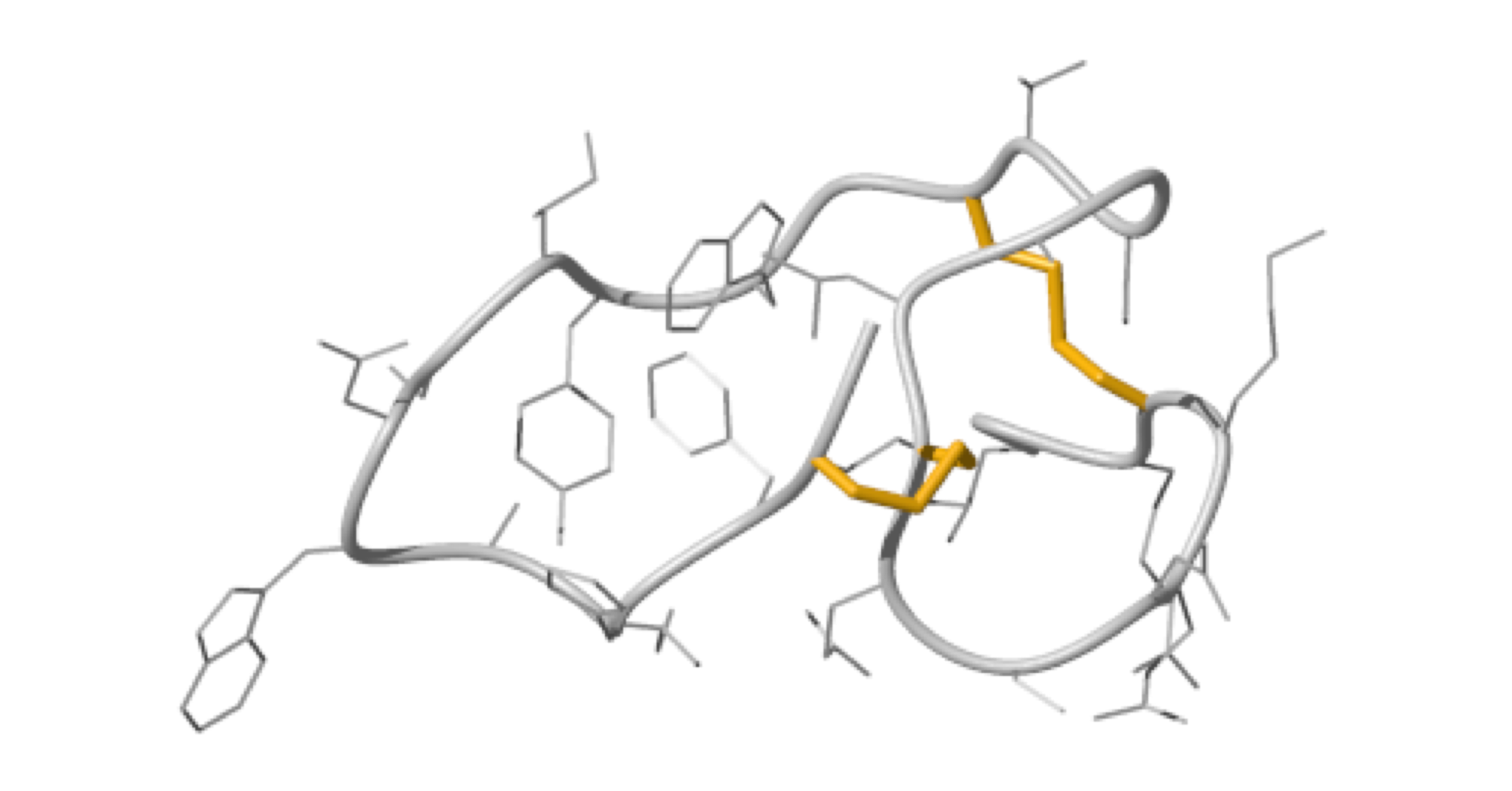 |
| Zoom region of the DLL-Rib peptide [1H-13C]-HSQC. Cystein's beta-carbons are highlighted in green | Ribbon 3D structure of the DLL-Rib peptide. Disulfide bridges are coloured in orange |
External Contact: U. Haberkorn (University of Heidelberg, Germany)
Zoller F, Markert A, Barthe P, Hebling U, Altmann A, Lindner T, Mier W & Haberkorn U. A disulfide-constrained miniprotein with striking tumor-binding specificity developed by ribosome display. Angew Chem Int Ed Engl. 2013 Nov 4;52(45):11760-4. doi: 10.1002/anie.201304603.
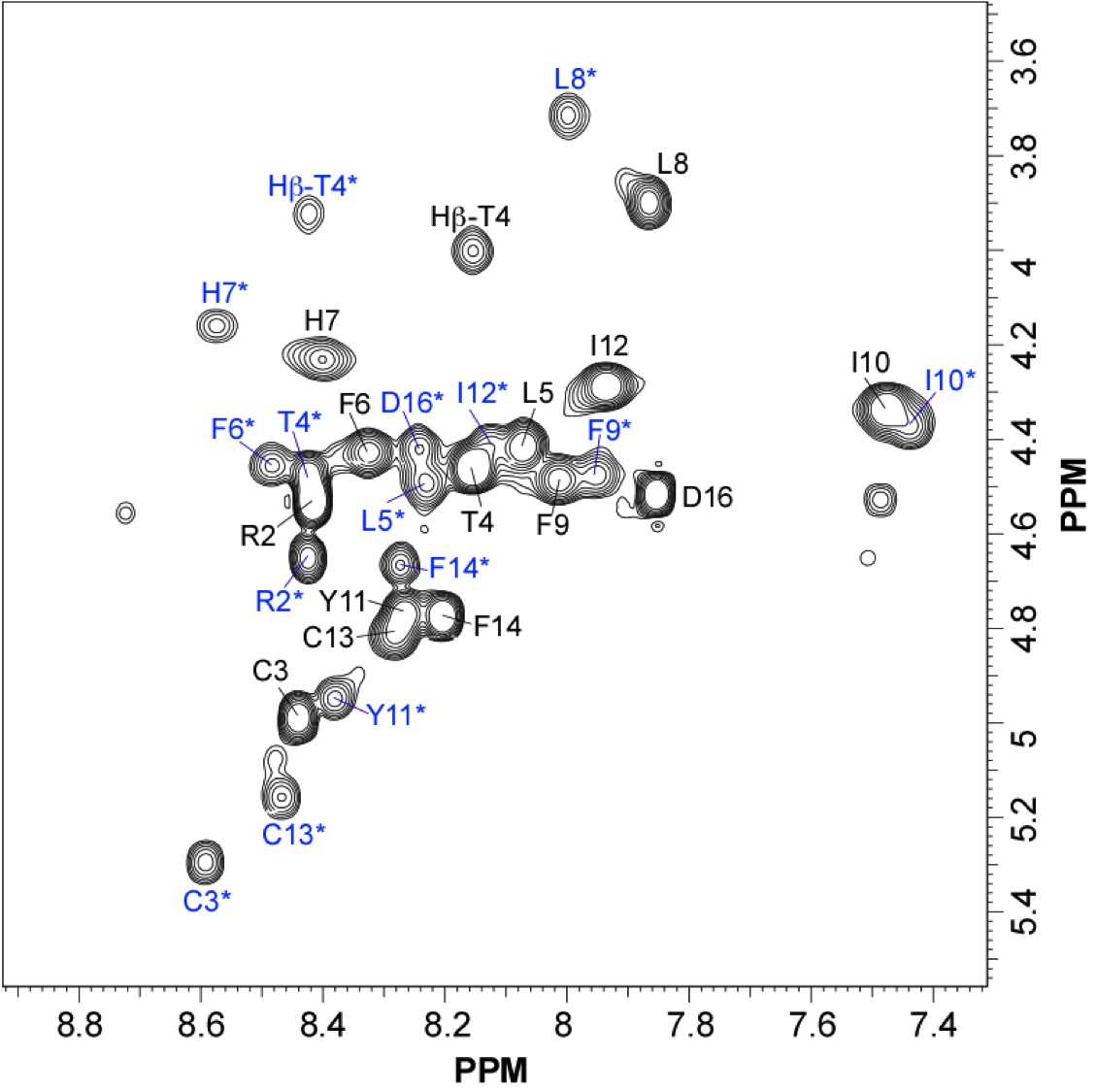
NMR structure of Bc28.1
NMR Platform Staff: Y. Yang & C. Roumestand
Protein Size : 223 a.a. 15N and 13C labeled
Babesia canis is the agent of the canine babesiosis in Europe. The major merozoite surface antigens of Babesia canis have been described as a 28-kDa membrane protein family, anchored at the surface of the merozoite. The resolution of the structure of Bc28.1 (223 a.a) represents a milestone for the characterization of the parasite erythrocyte binding and its interaction with the host immune system.
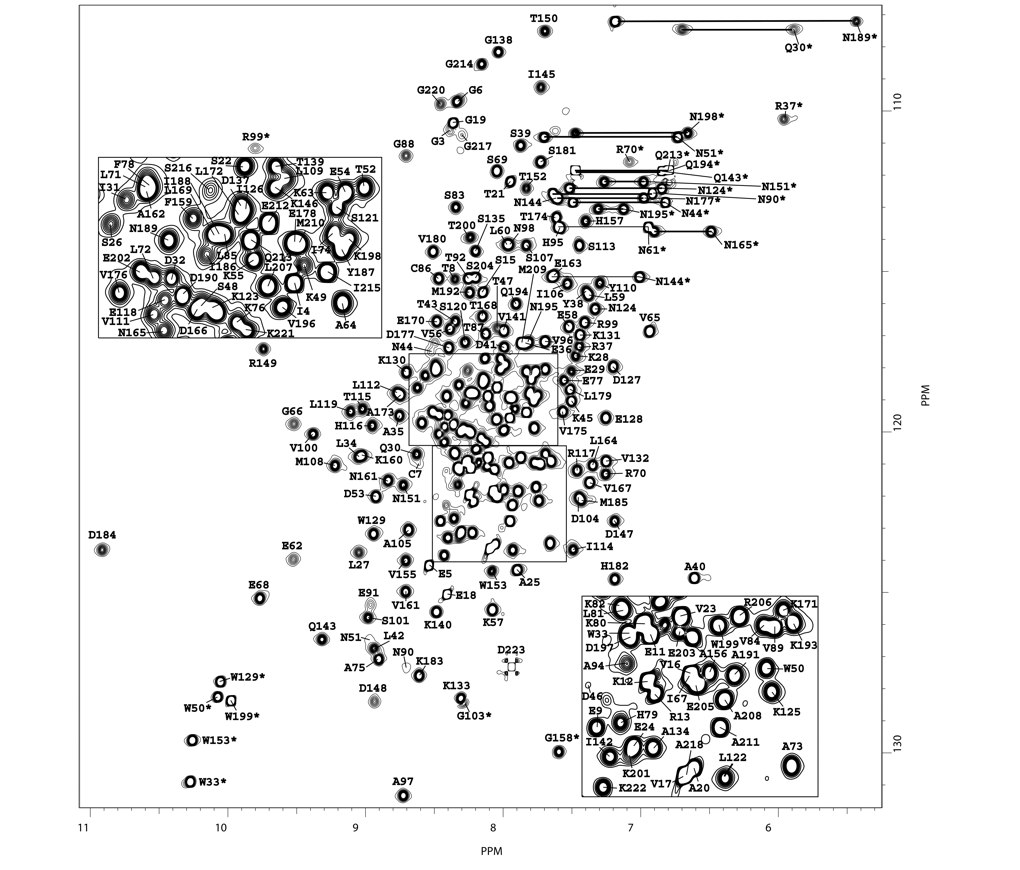 |
 |
| Left Panel : [1H-15N]-HSQC and zooms of lightly crowded areas |
External Contact : Stéphane Delbecq (EA4558, Vaccination Antiparasitaire, Montpellier)
Yang YS, Murciano B, Moubri K, Cibrelus P, Schetters T, Gorenflot A, Delbecq S & Roumestand C. Structural and functional characterization of Bc28.1, major erythrocyte-binding protein from Babesia canis merozoite surface, J Biol Chem. 2012 Mar 16;287(12):9495-508. doi: 10.1074/jbc.M111.260745.
NMR structure of a tumor-targeting chimeric protein
NMR Platform Staff : Philippe Barthe
We used the disulfide-stabilized miniprotein Min-23 as a molecular scaffold in a phage display approach to generate a combinatorial library for the identification of new affinity functions against the angiogenesis marker delta-like ligand 4 (Dll4). The grafting of the binding domain from miniprotein Min-23 into the sunflower trypsin inhibitor (SFTI-I) peptide scaffold preserved its in vitro and in vivo binding specificity and proteolytic stability. Both the Dll4-binding specificity and the tumor-targeting capability of the rationally designed miniprotein (SFMIN3) were confirmed in vitro and in vivo.
 |
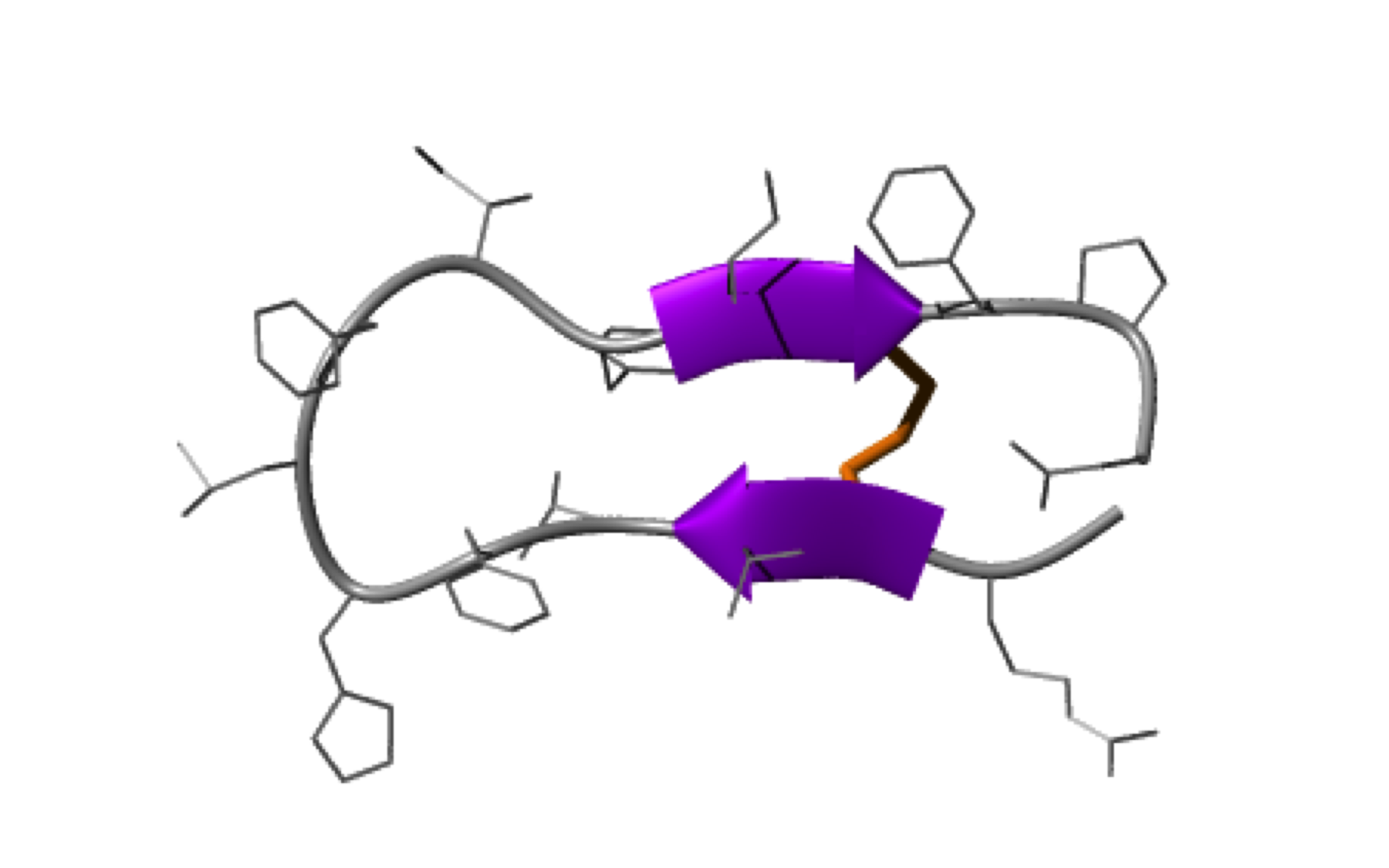 |
| Fingerprint TOCSY of the major (black) and minor (blue*) forms of SFMIN3 peptide | Ribbon 3D structure of the SFMIN3 peptide. Beta-sheet is represented by purple arrows, disulfide bridge is coloured in orange |
External Contact: U. Haberkorn (University of Heidelberg, Germany)
Zoller F, Markert A, Barthe P, Zhao W, Weichert W, Askoxylakis V, Altmann A, Mier W & Haberkorn U. Combination of phage display and molecular grafting generates highly specific tumor-targeting miniproteins. Angew Chem Int Ed Engl. 2012 Dec 21;51(52):13136-9. doi: 10.1002/anie.201203857.
2011
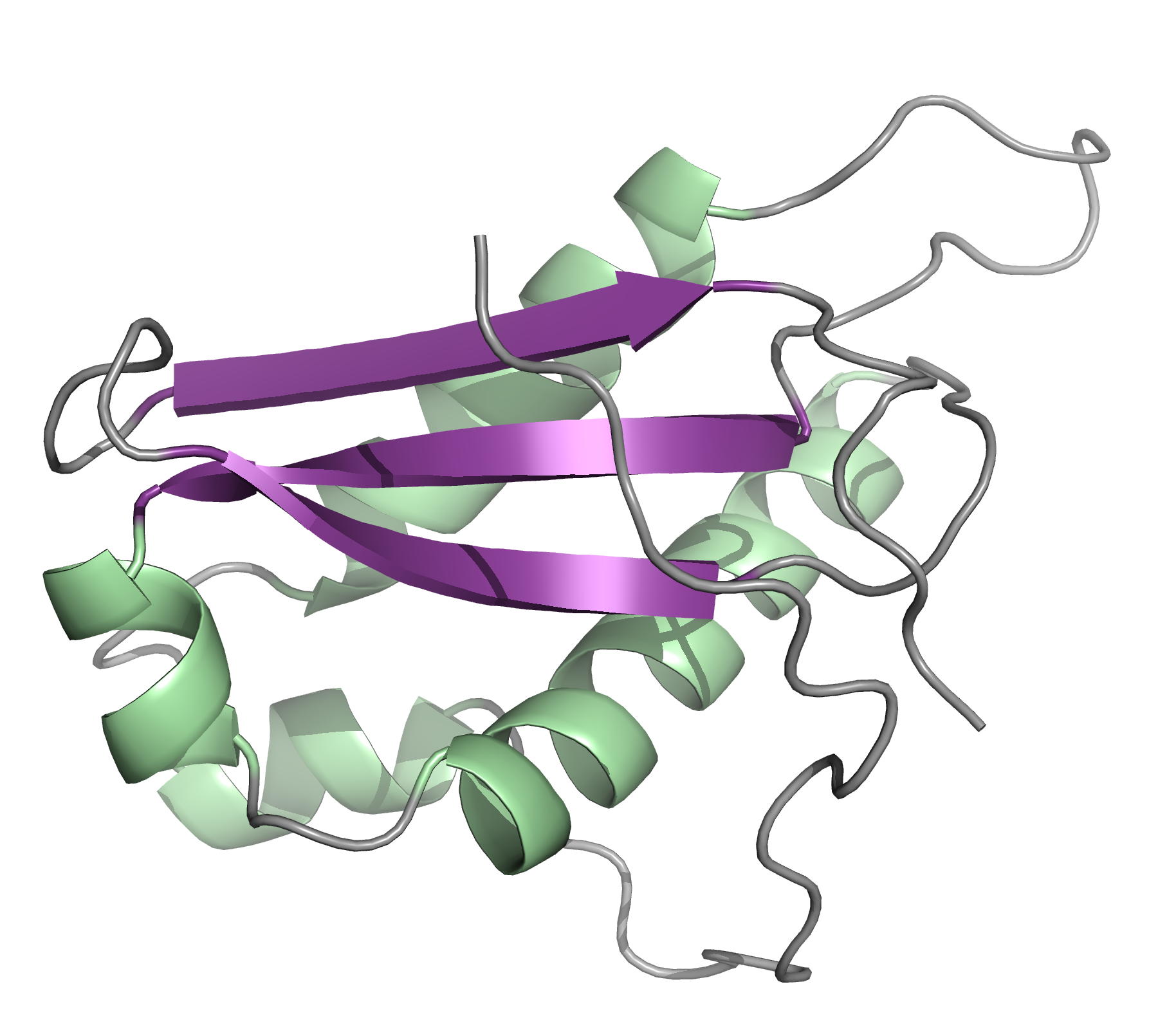
Solution structure of N- and C-terminal domains of Rv0020c from Mycobacterium tuberculosis
NMR Platform Staff : P. Barthe, A. Padilla & C. Roumestand
Protein Size : 132 a.a. and 100 a.a. 15N labeled
The protein Rv0020c from Mycobacterium tuberculosis, also called FhaA, is one of the major substrates of the essential Ser/Thr protein kinase PknB. The protein is composed of three domains. We solved the solution structure of both N- and C-terminal domains and demonstrated that the approximately 300 amino acids of the intermediate domain are not folded. We showed that the FHA domain of Rv0020c interacts with the phosphorylated juxtamembrane domain of PknB.
 |
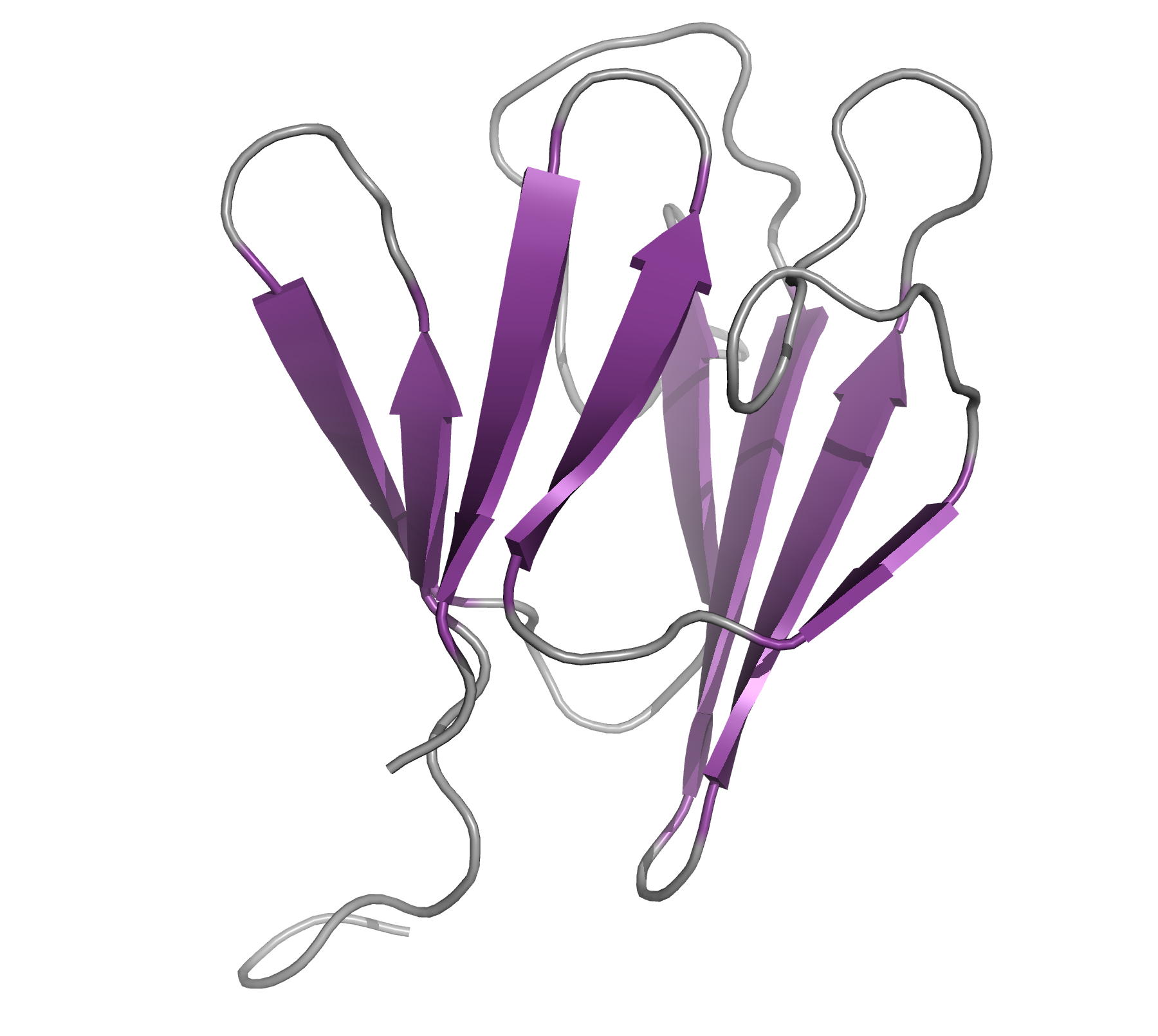 |
| Ribbon 3D structure of the Rv0020c_Nter | Ribbon 3D structure of the Rv0020c_Cter (FHA domain) |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Roumestand C, Leiba J, Galophe N, Margeat E, Padilla A, Bessin A, Barthe P, Molle V & Cohen-Gonsaud M. Structural insight into the Mycobacterium tuberculosis Rv0020c protein and its interaction with the PknB kinase. Structure. 2011 Oct 12;19(10):1525-34. doi: 10.1016/j.str.2011.07.011.
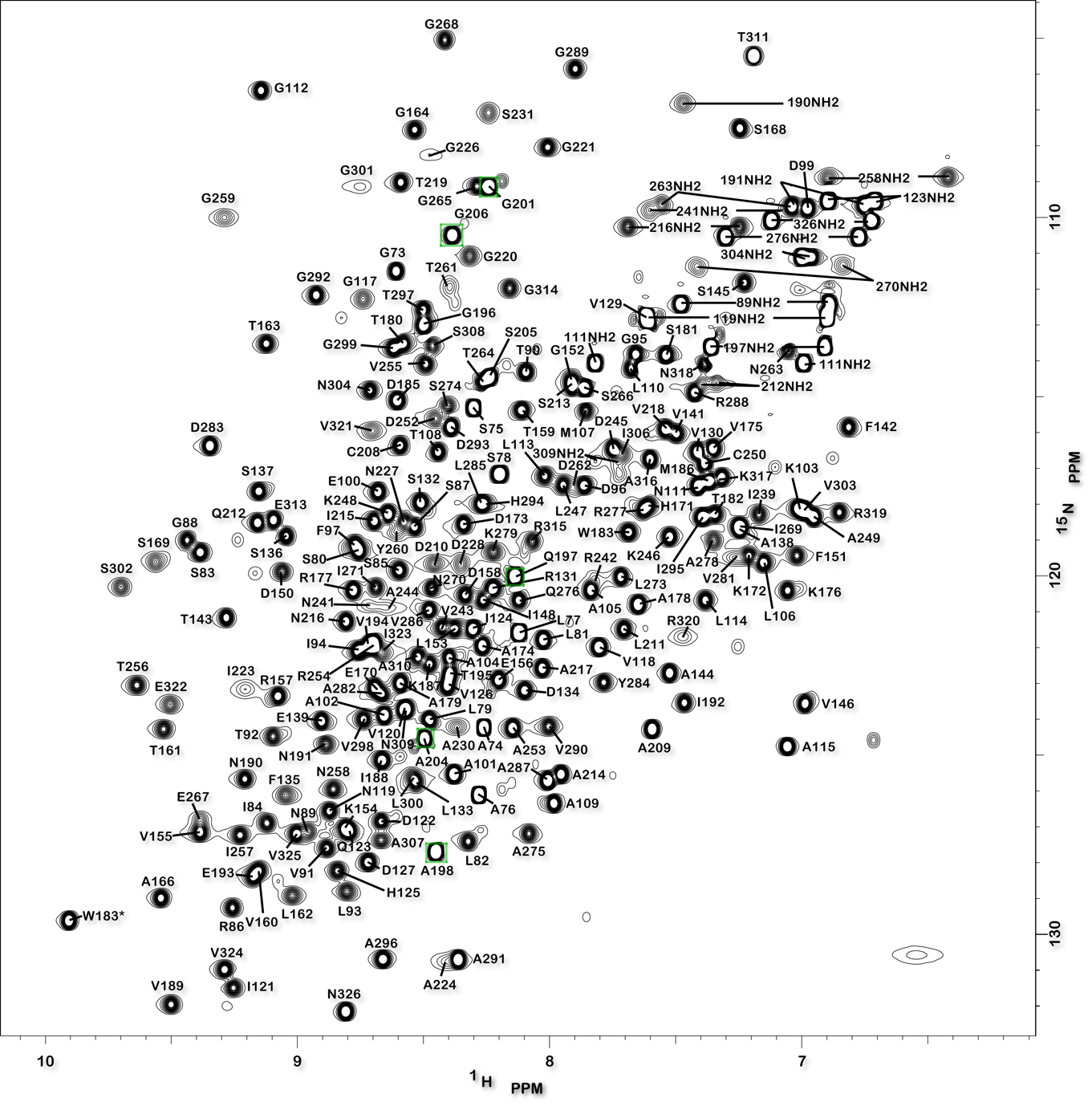
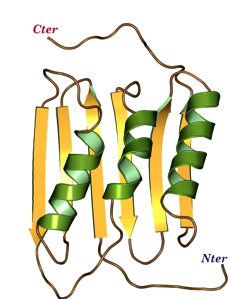
Structure of the Mycobacterium tuberculosis OmpATb
NMR Platform Staff: Y. Yang & C. Roumestand
Protein Size : 256 a.a. 15N and 13C labeled
The pore-forming outer membrane protein OmpATb from Mycobacterium tuberculosis is a virulence factor shown to be required for acid resistance in host phagosomes. NMR studies show that OmpATb is composed of two independent domains separated by a proline-rich hinge region. The results revealed the N-terminal domain of OmpATb can assemble into multiple oligomeric forms and is responsable to form channels in planar lipid bilayers.
 |
 |
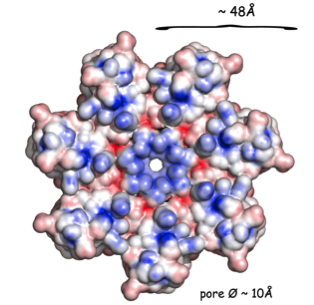 |
| [1H-15N]-HSQC spectrum 256 a.a. | N-terminal domain structure | HADDOCK model of the heptamer |
External Contact : Nathalie Saint (INSERM U1046, Montpellier)
Yang Y, Auguin D, Delbecq S, Dumas E, Molle G, Molle V, Roumestand C & Saint N. Structure of the Mycobacterium tuberculosis OmpATb protein: a model of an oligomeric channel in the mycobacterial cell wall. Proteins. 2011 Feb;79(2):645-61. doi: 10.1002/prot.22912.
2010
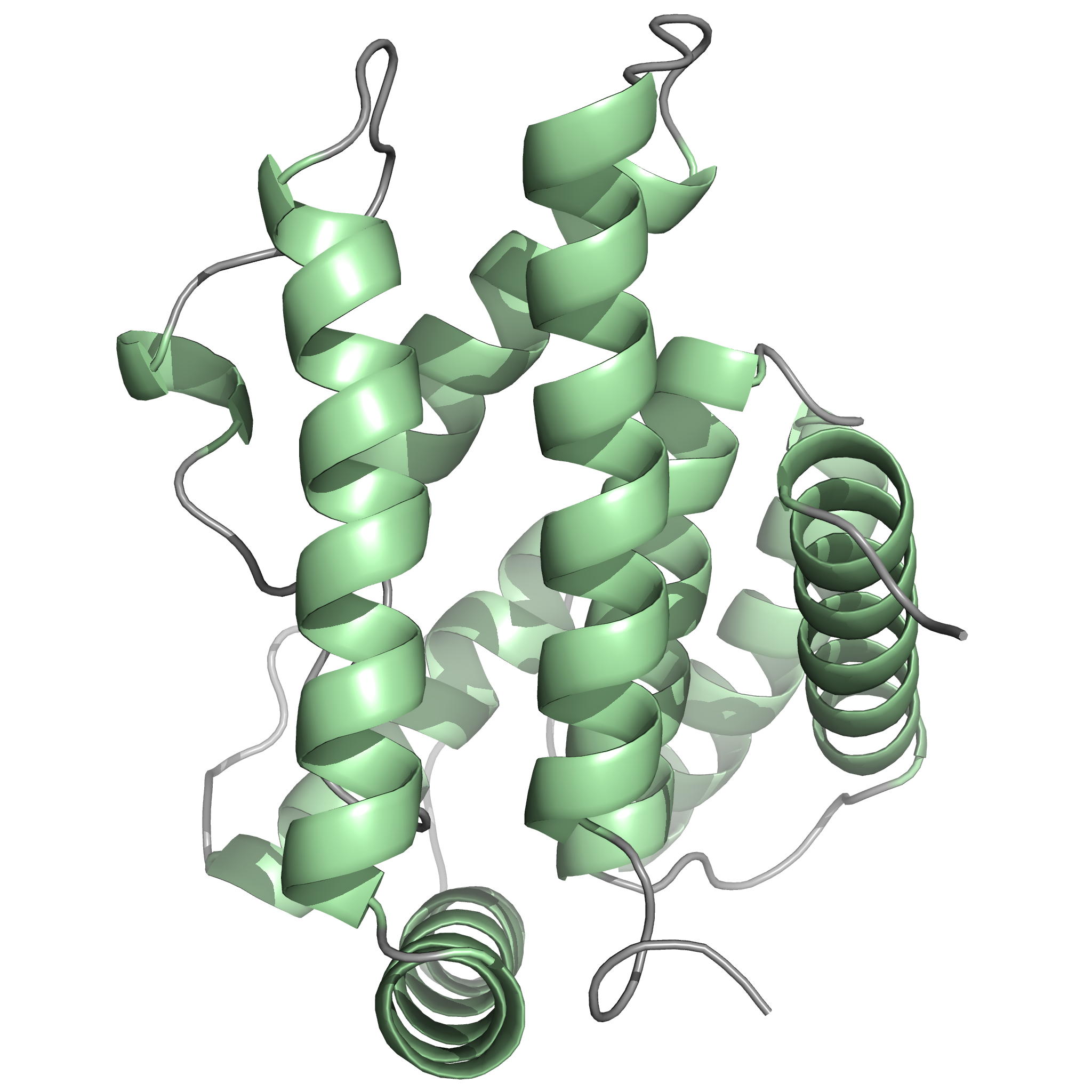
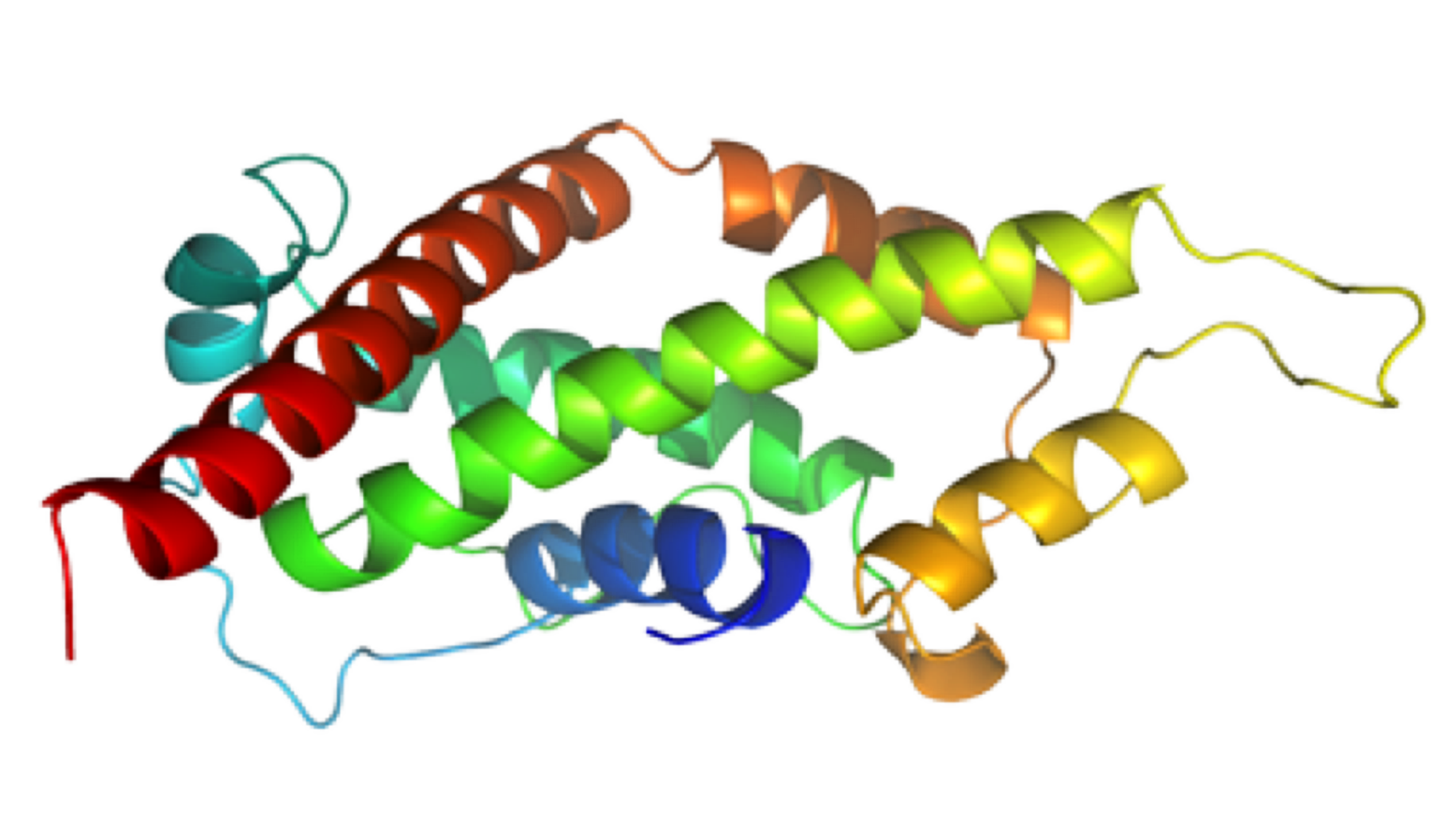
NMR Structure of PknB extracellular PASTA domain from Mycobacterium tuberculosis
NMR Platform Staff : P. Barthe & C. Roumestand
Protein Size : 272 a.a. 15N and 13C labeled
PknB is a transmembrane Ser/Thr protein kinase essential for Mycobacterium tuberculosis growth. The kinase possesses an extracellular region composed of a repetition of PASTA domains, believed to bind peptidoglycan fragments that might act as a signaling molecule. We report here the first solution structure of this extracellular region composed of 272 amino acids.
 |
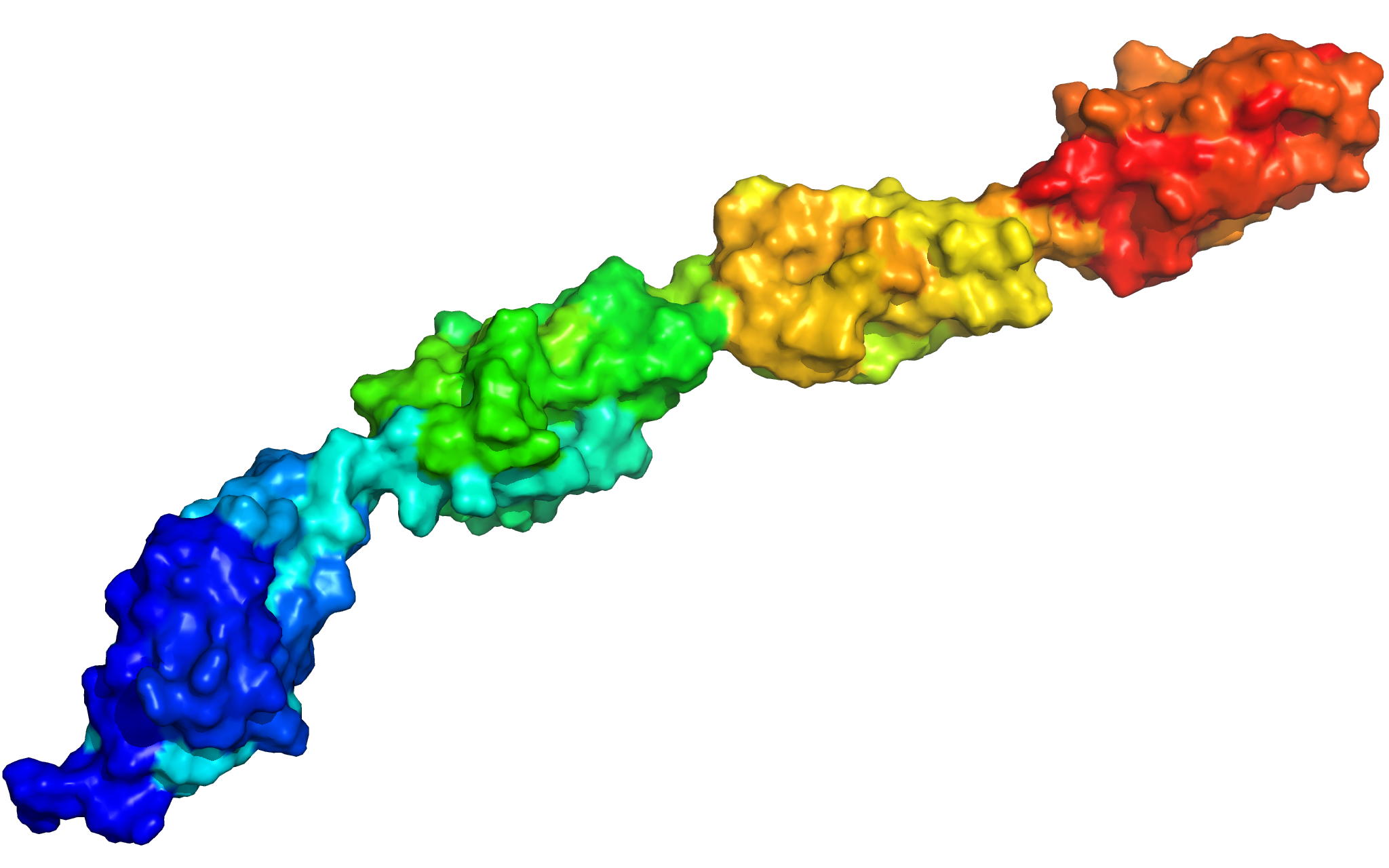 |
| Cover of the Structure journal | NMR solution structure of the extracellular PASTA domain protein from Mtb colored in rainbow (from blue N-terminus to red C-terminus) |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Barthe P, Mukamolova G, Roumestand C & Cohen-Gonsaud M. The strucutre of PknB extracellular PASTA domain from Mycobacterium tuberculosis suggests a ligand-dependent kinase activation. Structure. 2010 May 12;18(5):606-15. doi: 10.1016/j.str.2010.02.013.
2009
Solution structure of the unphosphorylated and phosphorylated isoforms of OdhI
NMR Platform Staff : P. Barthe & C. Roumestand
Protein Size : 143 a.a. 15N and 13C labeled
The OdhI protein is key regulator of the TCA cycle in Corynebacterium glutamicum. The unphosphorylated form of OdhI inhibits the OdhA protein, a key enzyme of the TCA cycle, whereas the phosphorylated form is inactive. In this study, we solved the solution structure of the unphosphorylated and phosphorylated isoforms of the protein. We observed a major conformational change between the two forms corresponding to a new autoinhibition mechanism described for a FHA domain protein.
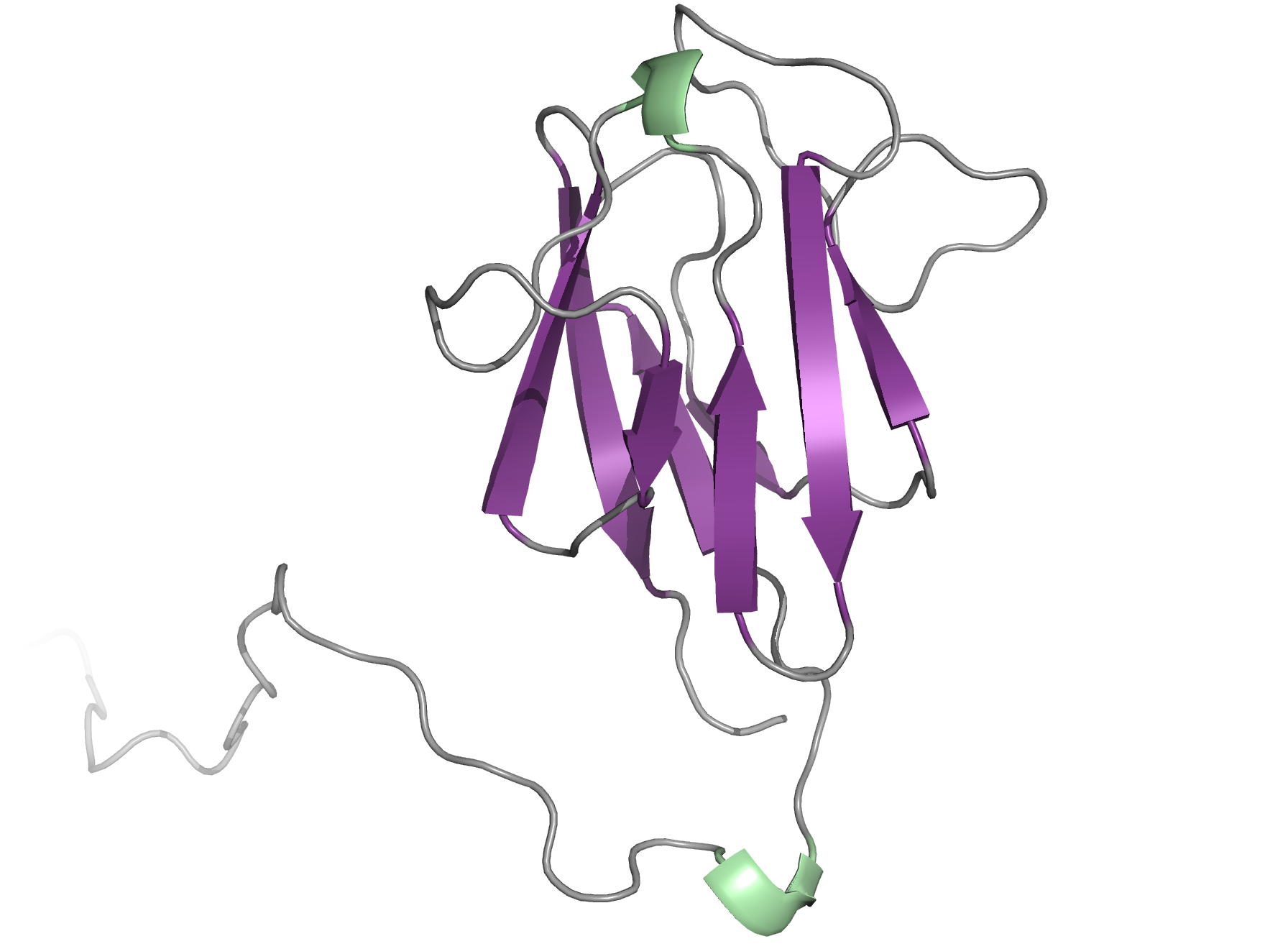 |
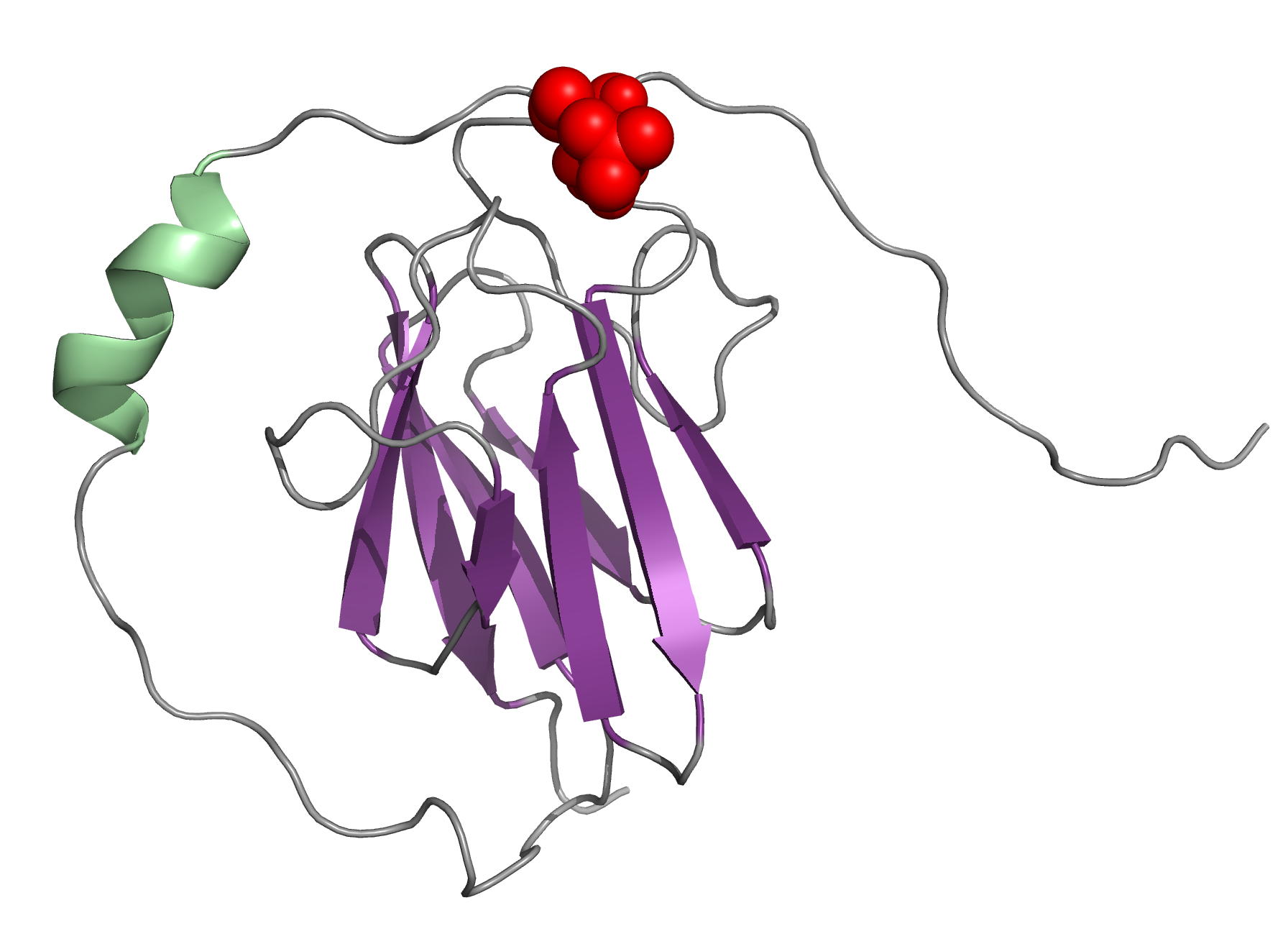 |
| NMR structure of the unphosphorylated-OdhI protein | NMR structure of the phosphorylated-OdhI protein |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Barthe P, Roumestand C, Canova M, Kremer L, Hurard C, Molle V & Cohen-Gonsaud M. Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure. 2009 Apr 15;17(4):568-78. doi: 10.1016/j.str.2009.02.012.
Solution structure of the Mycobacterium tuberculosis Rv2175c
NMR Platform Staff : P. Barthe & C. Roumestand
Protein Size : 146 a.a. 15N and 13C labeled
Rv2175c, a M. tuberculosis protein, is identified as a substrate of the PknL kinase. We solved the NMR solution structure of Rv2175c indicative of a DNA-binding protein. Mass spectrometry analyses identified Thr9 as the unique phosphoacceptor. The DNA-binding activity was completely abrogated in a Rv2175c_T9D mutant, designed to mimic constitutive phosphorylation, but not in a mutant lacking the first 13 residues. This implies that the function of the N-terminal extension is to provide a phosphoacceptor (Thr9), which, following phosphorylation, negatively regulates the Rv2175c DNA-binding activity.
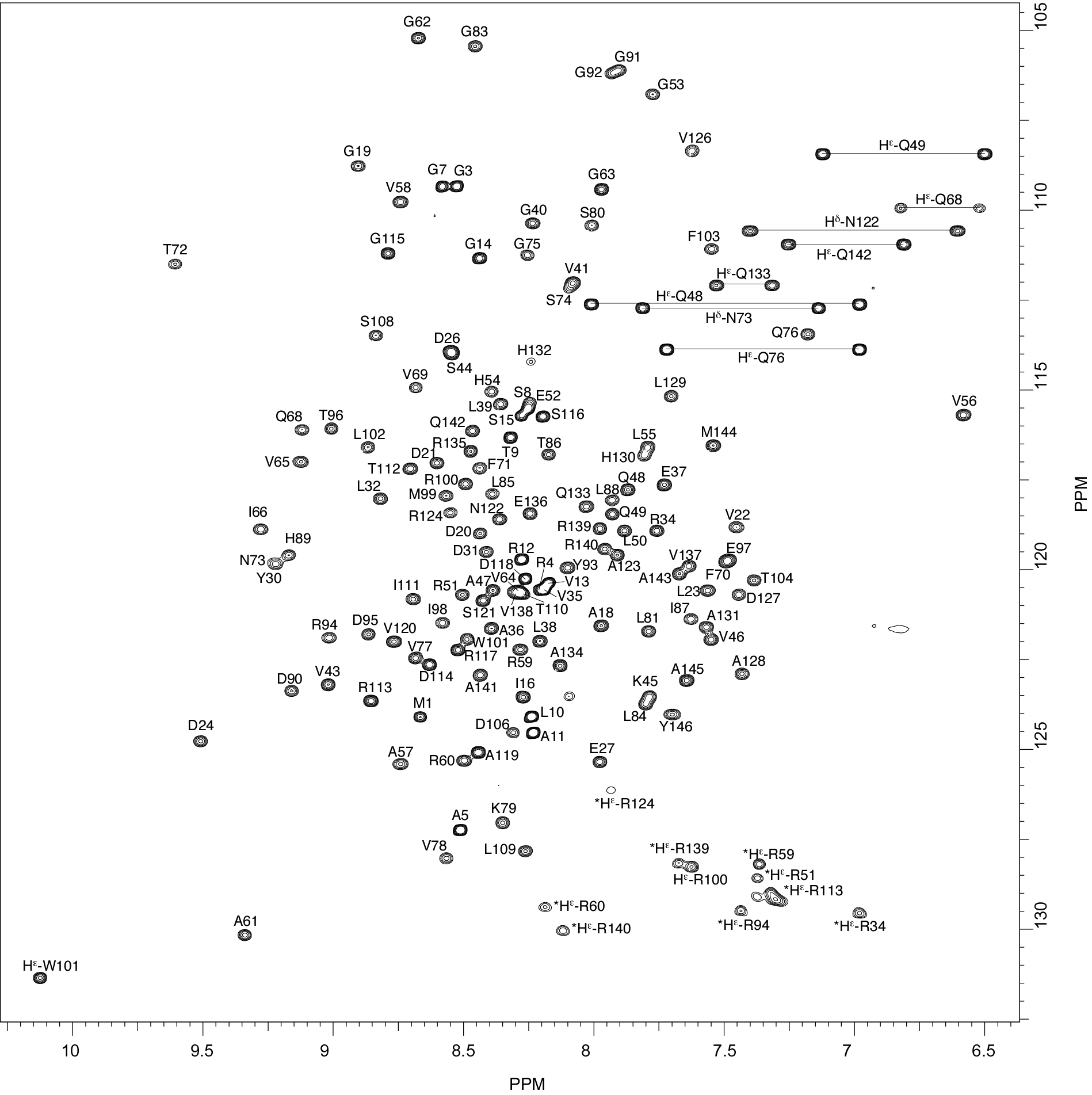 |
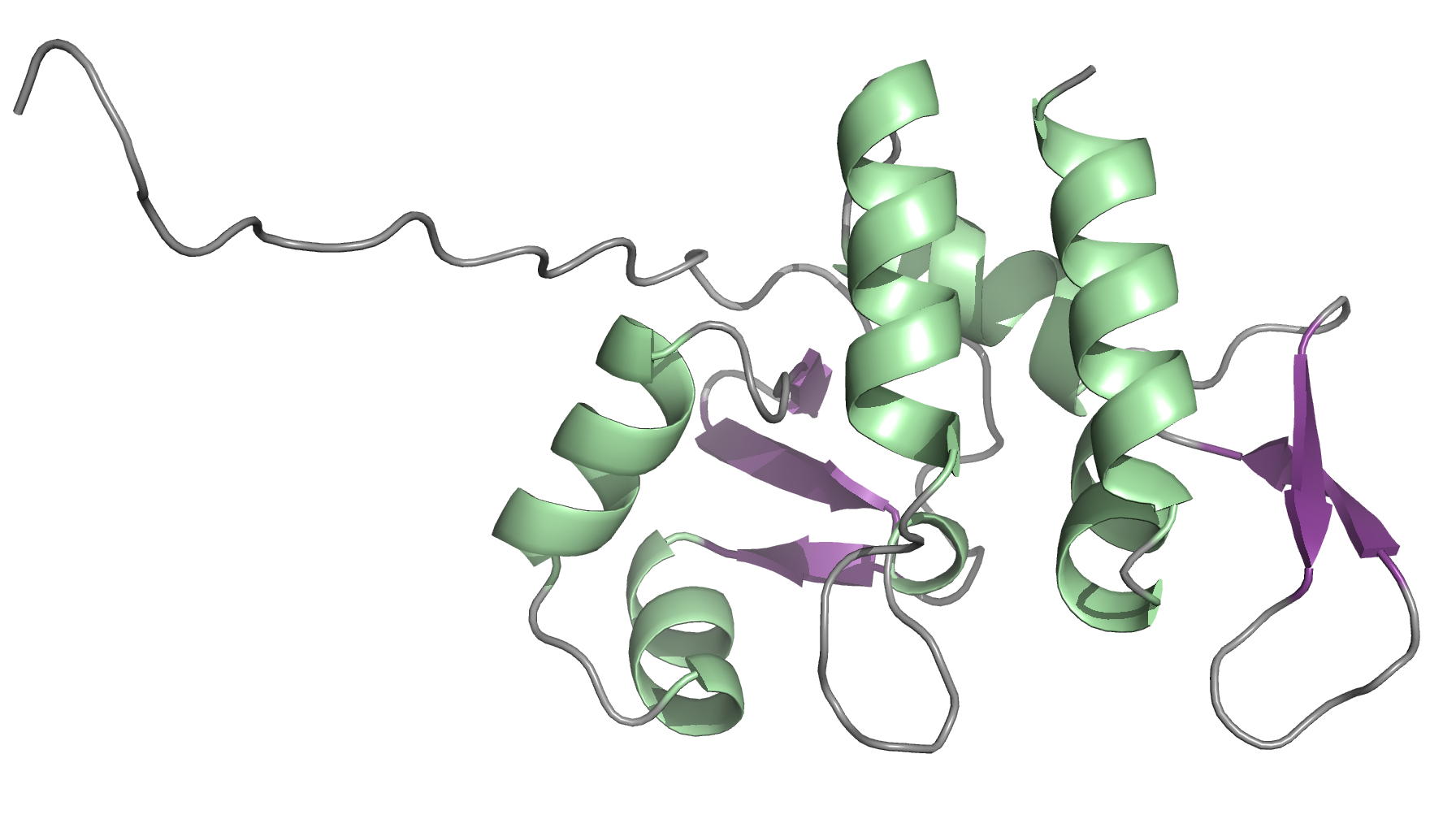 |
| [1H-15N]-HSQC of Rv2175c | Ribbon 3D structure of the Rv2175c protein |
Internal Contact: M. Cohen-Gonsaud (Centre de Biochimie Structurale, Montpellier)
Cohen-Gonsaud M, Barthe P, Canova M, Stagier-Simon C, Kremer L, Roumestand C & Molle V. The Mycobacterium tuberculosis Ser/Thr kinase substrate Rv2175c is a DNA-binding protein regulated by phosphorylation. J Biol Chem. 2009 Jul 17;284(29):19290-300. doi: 10.1074/jbc.M109.019653.
NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: model of the possible lipid A-binding site.
NMR Platform staff : Y. Yang & A. Padilla
Protein Size : 98 a.a. unlabeled
The anti-lipopolysaccharide factor ALF-Pm3 is a 98-residue protein identified in hemocytes from the black tiger shrimp Penaeus monodon. On the basis of the 3D structure and structural similarities to the FhuA/LPS complex, we designed an original model of the possible lipid A-binding site of ALF-Pm3. Delineating lipid A-binding site of ALFs will help go further in the de novo design of new antibacterial or LPS-neutralizing drugs.
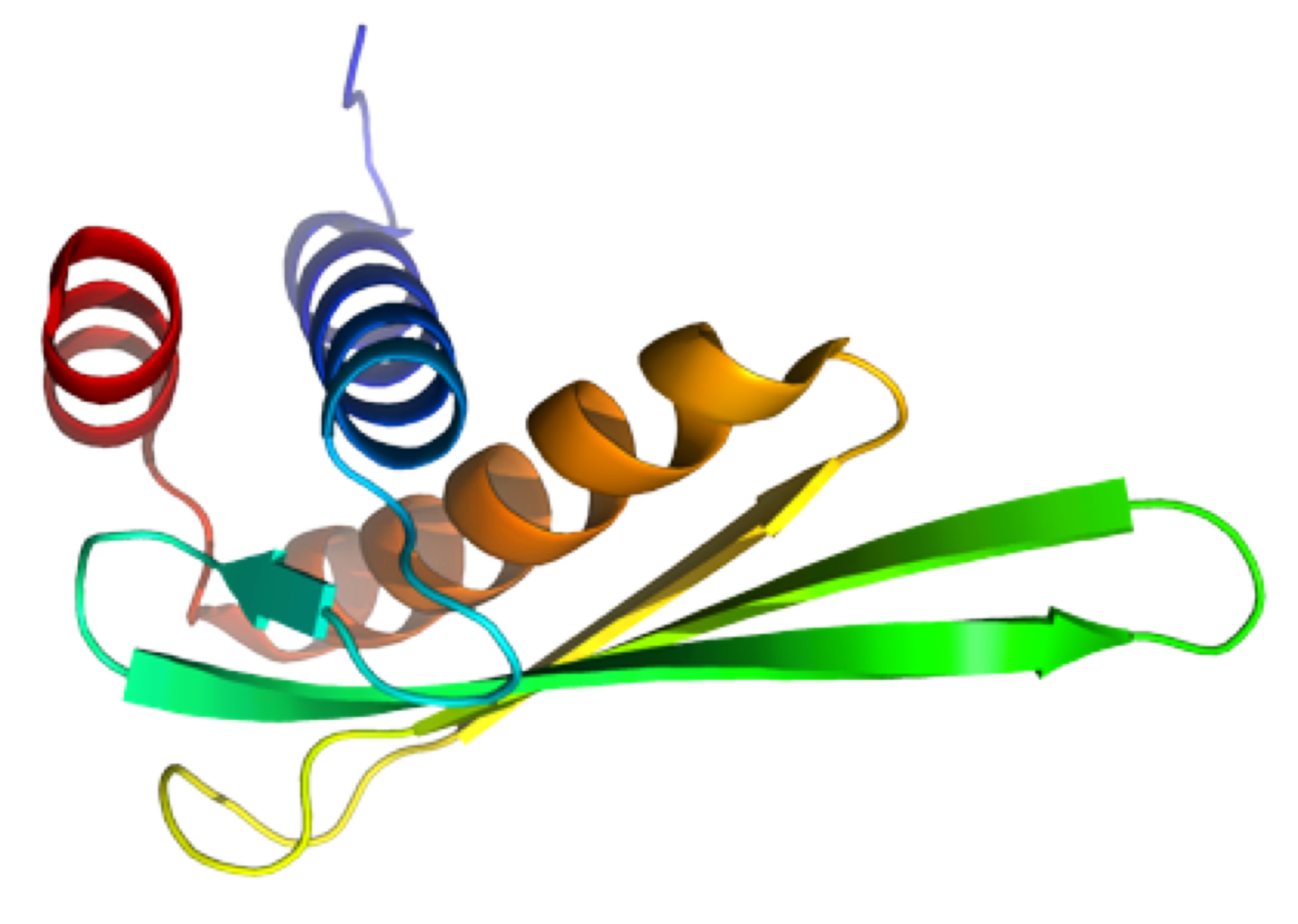
External Contact : Evelyne BACHERE (IFREMER-CNRS-Université Montpellier, FRE 2626)
Yang Y, Boze H, Chemardin P, Padilla A, Moulin G, Tassanakajon A, Pugniere M, Roquet F, Destoumieux-Garzon D, Gueguen Y, Bachere E & Aumelas A. NMR structure of rALF-Pm3, an anti-lipopolysaccharide factor from shrimp: model of the possible lipid A-binding site, Biopolymers. 2009 Mar;91(3):207-20. doi: 10.1002/bip.21119.
NMR structure of the mammalian deoxynucleotide N-hydrolase Rcl protein
NMR Platform Staff : Y. Yang & A. Padilla (Pipeline to the CBS RX Platform)
Protein Size : 2*150 a.a. 15N and 13C labeled
The gene Rcl encodes a deoxynucleoside 5′-monophosphate N-glycosidase that catalyzes the hydrolysis of the N-glycosidic bond of the nucleotide to give deoxyribose 5-phosphate and a nucleobase. The NMR study reveal an important ligand-induced stabilization of the dimer structure of the enzyme which can be used as a new and attractive therapeutic target.
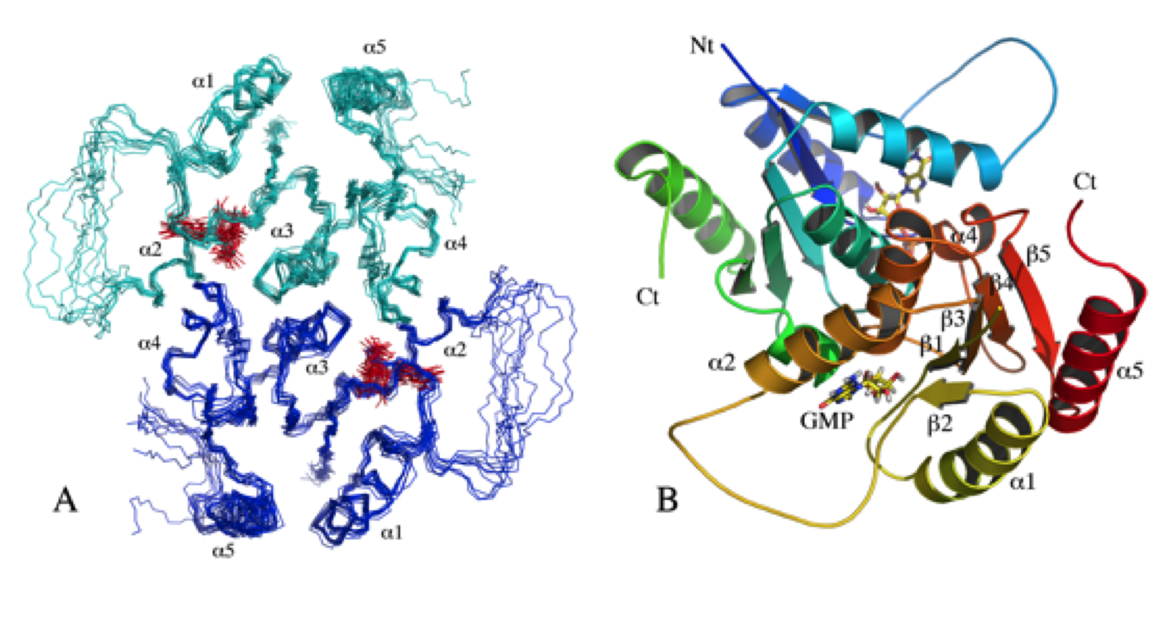 |
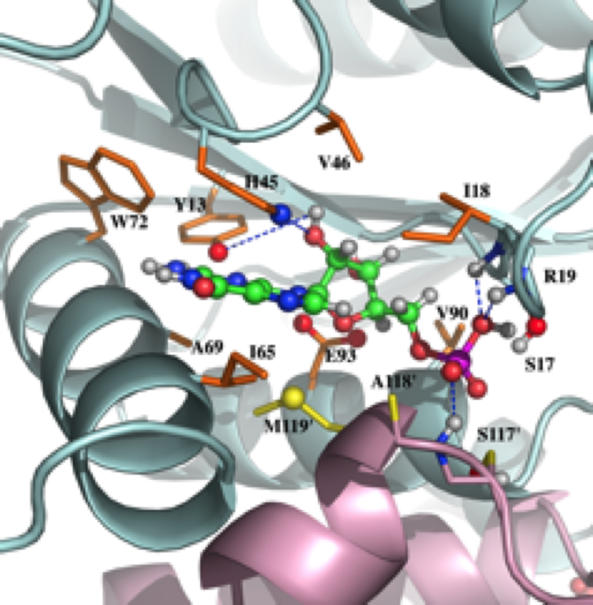 |
External Contact : P. Alexandre Kaminski (URA CNRS 2128, Institut Pasteur, 75724 Paris)
Yang Y, Padilla A, Zhang C, Labesse G & Kaminski P. Structural characterization of the mammalian deoxynucleotide N-hydrolase Rcl and its stabilizing interactions with two inhibitors, J Mol Biol. 2009 Dec 4;394(3):435-47. doi: 10.1016/j.jmb.2009.10.004.

