Speech can propagate pathogens

Speech is a potent route for viral transmission in the COVID-19 pandemic. Informed mitigation strategies are difficult to develop since no aerosolization mechanism has been visualized yet in the oral cavity nor has the relationship of speech to the exhaled flow been documented. In two recent studies, Manouk Abkarian together with Prof. Howard A. Stone from Princeton University in the USA have explored with high-speed imaging how phonation of common stop-consonants like 'P' or 'B', form and extend salivary filaments in a few milliseconds as moist lips open or when the tongue separates from the teeth. Both saliva viscoelasticity and airflow associated with the plosion of stop-consonants are essential for stabilizing and subsequently forming centimeter-scale thin filaments, tens of microns in diameter, that break into speech droplets [1] (see Figure A).
In collaboration with Simon Mendez from the university of Montpellier, these researchers showed that these plosive consonants induce starting jets and vortex rings that drive meter-long transport of exhaled air, tying this drop-formation mechanism to transport associated with speech [2]; the transport features, including phonetics, are demonstrated using order-of-magnitude estimates, numerical simulations, and laboratory experiments (see Figure B). These authors believe that these works will inform thinking about the role of ventilation, aerosol transport in disease transmission for humans and other animals, and yield a better understanding of ''aerophonetics.''
This research is being continued with the Metropolitan Opera Orchestra ("MET Orchestra") in New York, as part of a project to identify the safest conditions for continuing this prestigious orchestra's activity (Figure C).
[1] Speech can produce jet-like transport relevant to asymptomatic spreading of virus. M. Abkarian, S. Mendez, N. Xue, F. Yang, H. A. Stone, Proceedings of the National Academy of Sciences, le 25 septembre 2020 DOI : https://doi.org/10.1073/pnas.201215611710.1073/pnas.2012156117
[2] Stretching and break-up of saliva filaments during speech: a route for pathogen aerosolization and its potential mitigation. M. Abkarian, H. A. Stone, Physical Review Fluids, le 2 octobre 2020 DOI: https://doi.org/10.1103/PhysRevFluids.5.102301
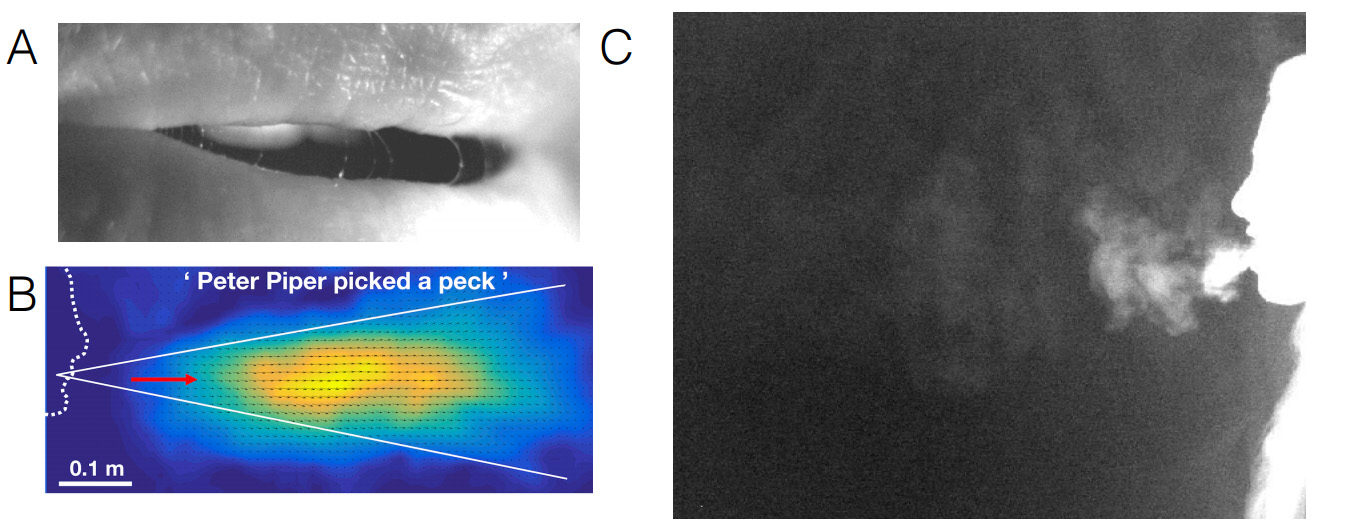
Figure (A) Close up of a mouth saying 'Pa'. (B) Average Flow Velocity indicating a conical jet-like structure when saying 'Peter Piper picked a peck', (C) CO2 exhaled air flow from a Mezzo Soprano Singer singing 'Oror' an Armenian Lullaby.
External links