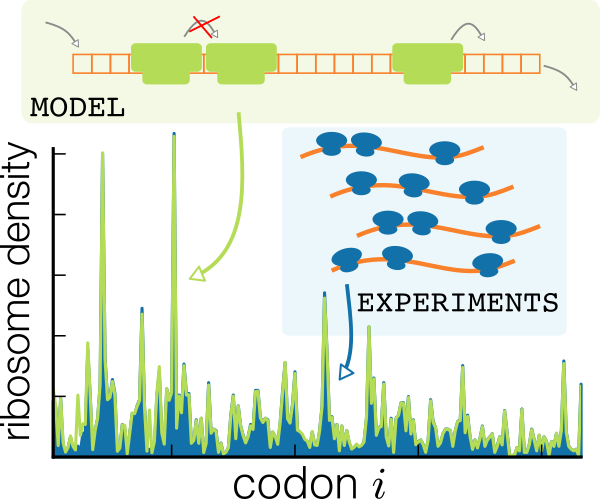
A non-equilibrium analysis model was developed for inferring the relative rates of translation initiation and elongation rates from ribosome profiling data.
One of the greatest challenges in modelling mRNA translation is to identify coding sequence features determining protein synthesis rates. Recent technical advances such as ribosome profiling allow probing translation at codon resolution, and make quantitative studies of transcript efficiency more accessible. We have developed a method called NEAR that integrates experimental ribosome profiling data and a well known non-equilibrium model of ribosome traffic. Our approach provides biological insights of traffic control and infers the kinetics of ribosomes on each transcript. When examining ribosome profiling in yeast, we observe that translation initiation and elongation are close to their optima and traffic is minimized at the beginning of the transcript to favour ribosome recruitment. Our work provides new measures of translation initiation and elongation efficiencies, emphasizing the importance of rating these two stages of translation separately.